There are generations of approaches. Often limited to the knowledge available at the time
Generation #1: Eat Fermented Foods as a Cure All
This dates back millennium in the east and the west. It helps some, and thus is validated as working (for some at least). For example, Garum in ancient Greece
Generation #2: Yogurt and Probiotics
In western culture, The Russian biologist and Nobel laureate Ilya Mechnikov, from the Institut Pasteur in Paris, was influenced by Grigorov’s work and hypothesized that regular consumption of yogurt was responsible for the unusually long lifespans of Bulgarian peasants.[25] Believing Lactobacillus to be essential for good health, Mechnikov worked to popularize yogurt as a foodstuff throughout Europe. [Wikipedia]
Generation #3: Bacteria Shifts
There are several generation of technology involved here.
“A significant difference in gut microbial composition between SARS-CoV-2 positive and negative samples was observed, with Klebsiella and Agathobacter being enriched in the positive cohort.”
The gut microbiome of COVID-19 recovered patients returns to uninfected status in a minority-dominated United States cohort [2021]
These studies indicates an increase or decrease in the average for populations. There is no thresholds where the odds change nor relative magnitude. This is further complicated by non-replication by other researchers — the reason is often because on non-standardization of microbiome analysis
Generation #4: Lab Specific Shifts with critical levels and contributions
Using large dataset and techniques such as those described in Symptoms with Ability to Predict from Microbiome Results. We have the ability to set threshold and determine the relative importance. The table below is for Long COVID based on one lab’s pipeline. We can easy see the pattern — often, it is a relatively rare bacteria(low prevalence) that is seen in significant levels in Long COVID patients
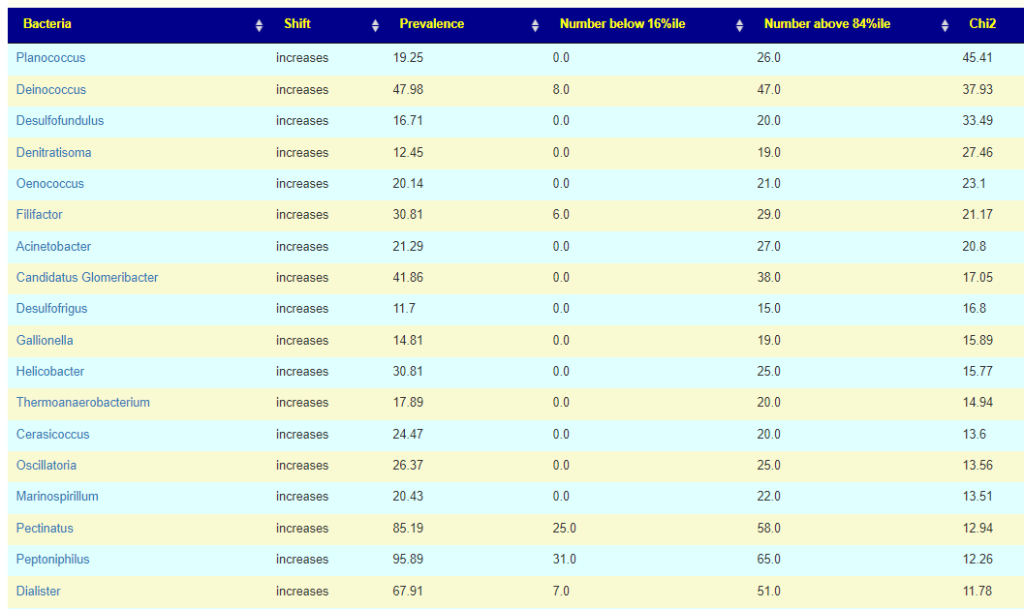
This allows identification of the genus (or other ranks) that may be ascribe to the condition if over the 84%ile. It also allows the relative importance of each to be evaluated since there may be multiple targeted bacteria. Chi2 value is a reasonable proxy for importance.
Moving up the taxonomical rank, we see at the ORDER level that one order is really significant.
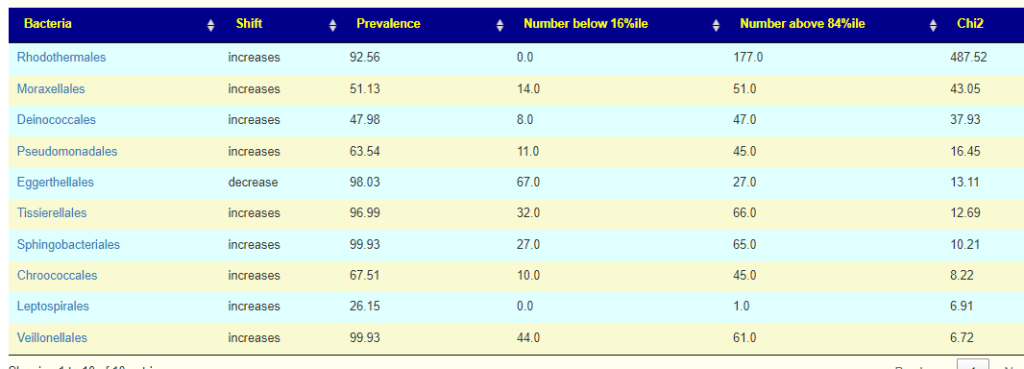
Bottom Line
IMHO, this last method allows superior identification of bacteria involved with conditions and symptoms using two simple cutoff points: <= 16%ile and >=84%ile. Other cutoff points are possible,
We can then look at a patient’s microbiome (assuming suitable lab-pipeline) and identify with statistical accuracy which bacteria are involved. Not only can we identify the bacteria — we can determine the relative importance of each bacteria.
Recent Comments