In a clinical setting, a practitioner may conceptually believe that a patient would benefit from a probiotic. The problem is which one(s). Often the advice is a generic “take a good probiotic”; a suggestion bordering on magical thinking.
Video version below.
Level 1: Using Published Studies
In general, published studies use specific strains of probiotics. Those strains may not be readily available. Often, the suggestion would be to take the same species (with fingers crossed).
For those that wish to avoid this wishful thinking, we have a page listing Research Probiotics available Retail. This allows you to do a quick search. For example, for ADHD we have just two strains listed as shown below. For some conditions, nothing will be found. These are links to studies or reviews that need to be reviewed by the practitioner.
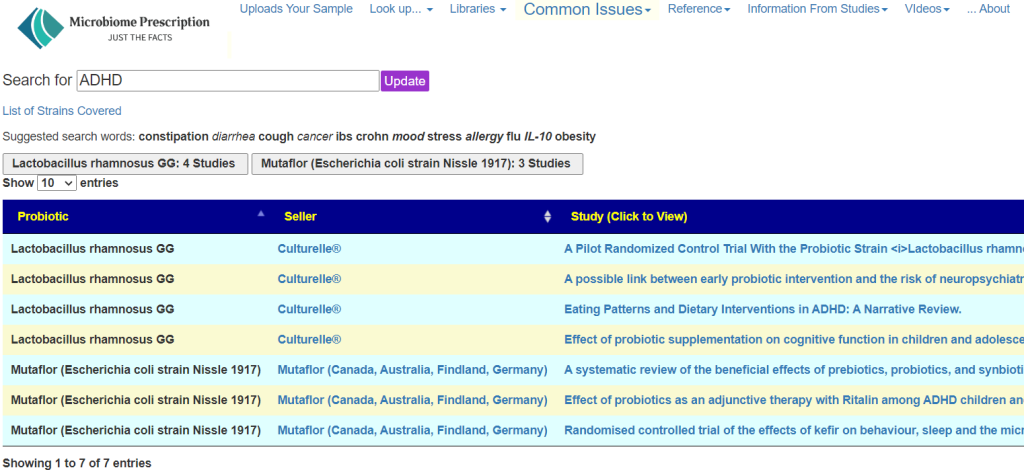
The basic issue is a lack of studies. Comparison studies are usually non-existant.
Level 2: Identifying cause of condition(s) and targeting taxa
Often this is done by using microbiome analysis looking for abnormal levels of bacteria and seeing what will alter them. For example, multiple studies report low levels of Faecalibacterium and high levels of Bifidobacterium for ADHD. As above, we have a search page that links to studies of the impact of different probiotics (and supplements) on each bacteria.
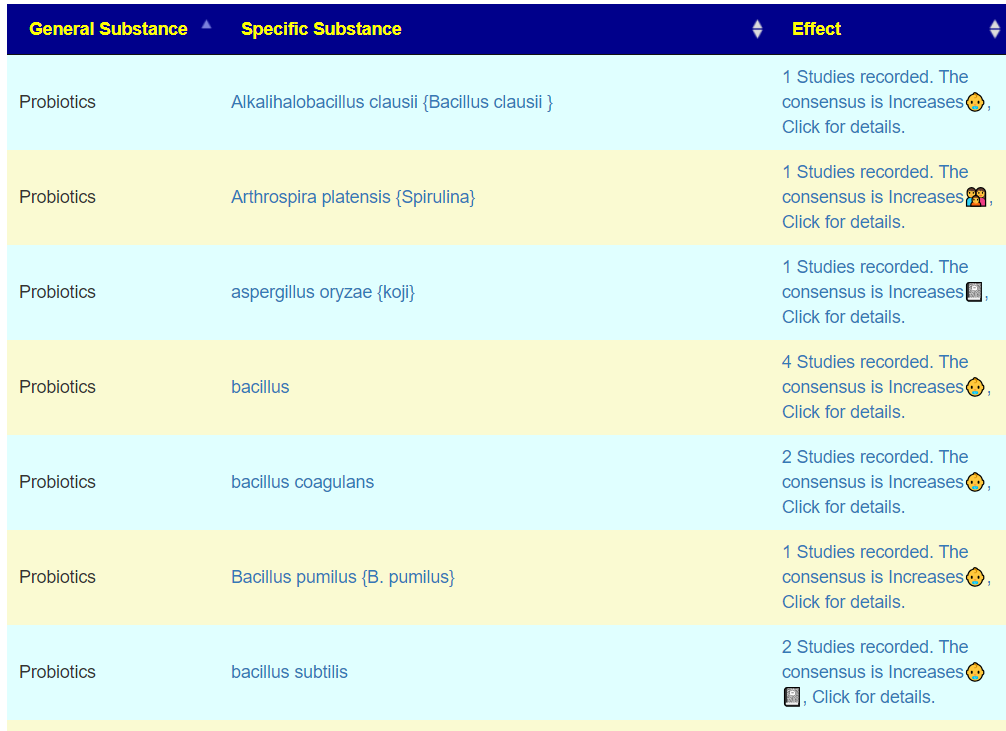
Level 3: Identifying cause using Enzymes and Metabolites
At this point we enter into the Citizen Science world at Microbiome Prescription. Thousands of people have uploaded their microbiome samples from a host of different providers and then annotated the samples with their symptoms and conditions. The data is at MicrobiomePrescription Citizen Science.
The chart below shows the process. The number of abnormal bacteria (too high or too low) is much larger than published studies — not unexpected given the much larger sample size.
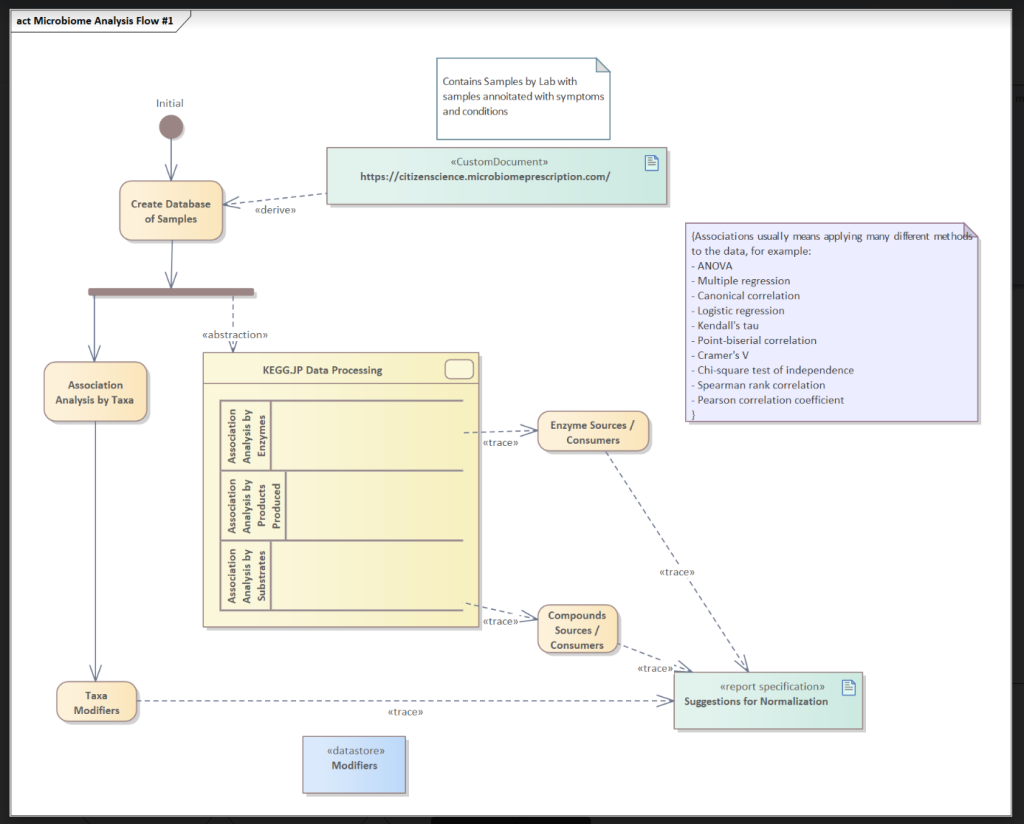
Abstraction
We take the microbiome data and transformed it with data from KEGG: Kyoto Encyclopedia of Genes and Genomes to get estimates of enzymes and metabolites or compounds. This data is processed thru a variety of methods to determine associations of the enzymes and metabolites to condition.
What we observe is that at the metabolite level we often have agreement across the three most common providers

At the enzyme level, we do not get this strong pattern
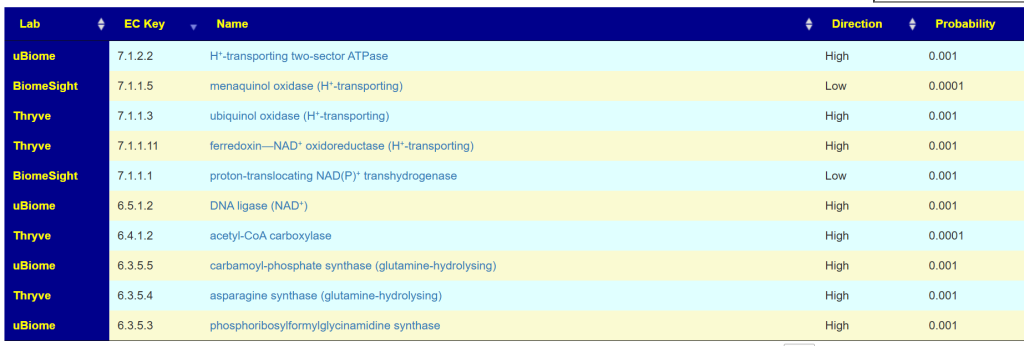
Nor do we get it by the bacteria associated.
Apparent Conclusion
The cause of the symptom or diagnosis appears to be an imbalance of the metabolites. Metabolites levels are the results of multiple bacteria and not a specific bacteria.
Monte Carlo Selection of Probiotics
As a proof of concept, I applied algorithms to the above with the following being the top items suggested (in descending priority). Play with it on Symptom Association Studies.
- Taxa Based — Select probiotics based on abnormal bacteria shifts
- Enzyme Based — Select probiotics based on enzymes that are deficient in the condition, but know to be produced by the probiotic
- Metabolite Based — Select probiotics based on metabolites that are deficient in the condition, which the probiotic impacts
| Taxa Based | Enzyme Based | Metabolite Based |
| clostridium butyricum ,Miya,Miyarisan Lentilactobacillus kefiri {Kefibios} bifidobacterium lactis,streptococcus thermophilus probiotic pediococcus acidilactic {RBB9 PEDIOCOCCUS ACIDILACTI} Bifidobacterium animalis Lacticaseibacillus paracasei shirota {Yakult} bifidobacterium infantis {B. infantis} lactobacillus helveticus {L. helveticus} Bifidobacterium animalis subsp. lactis {B. Lactis} lactobacillus reuteri bifidobacterium longum,lactobacillus helveticus Levilactobacillus brevis {L.brevis} Bacillus pumilus {B. pumilus} lactobacillus salivarius Lactobacillus Johnsonii {Lactobacillus Johnsonii} lactobacillus paracasei,lactobacillus acidophilus,bifidobacterium animalis lactobacillus paracasei Streptococcus faecalis, Clostridium butyricum, Bacillus mesentericus {Bio-three} Lentilactobacillus buchneri {Lactobacillus buchneri} Lactobacillus kefiranofaciens {Kefir Probiotic} bifidobacterium pseudocatenulatum li09,bifidobacterium catenulatum li10 mutaflor escherichia coli nissle 1917 enterococcus faecium (probiotic) Pediococcus pentosaceus lactobacillus helveticus,lactobacillus rhamnosus Bifidobacterium longum subsp. longum BB536 {BB536} lactobacillus plantarum,xylooligosaccharides, lactobacillus crispatus {L. Crispatus} Enterococcus faecium sf 68 {bioflorin} aspergillus oryzae {koji} lactobacillus casei Bifidobacterium breve {B. breve} Latilactobacillus sakei {Lactobacillus sakei} Arthrospira platensis {Spirulina} Brevibacillus laterosporus {B. laterosporus } Lactobacillus jensenii {L Jensenii} Escherichia coli cryodesiccata {colinfant probiotics} Finnish Probiotic {Valio Probiotic} Alkalihalobacillus clausii {Bacillus clausii } bifidobacterium bifidum bacillus subtilis,lactobacillus acidophilus Limosilactobacillus fermentum (probiotic) Bifidobacterium catenulatum subsp. catenulatum {Bifidobacterium catenulatum} Escherichia coli:DSM 16441-16448 {symbioflor-2} lactobacillus plantarum bacillus subtilis natto {B.natto} Lactiplantibacillus pentosus {L. pentosus} Bacillus amyloliquefaciens group {B. Amyloliquefaciens} Lactococcus lactis {Streptococcus lactis} Lactobacillus gasseri {L.gasseri} | Pseudomonas fluorescens Pseudomonas putida Escherichia coli Azospirillum lipoferum Azospirillum brasilense Cereibacter sphaeroides Rhodospirillum rubrum Streptomyces venezuelae Azotobacter vinelandii Rhodococcus rhodochrous Azotobacter chroococcum Pimelobacter simplex Acinetobacter calcoaceticus Priestia megaterium Streptomyces fradiae Brevibacillus brevis Bacillus thuringiensis Peribacillus simplex Paenibacillus polymyxa Bacillus subtilis Arthrobacter citreus Brevibacillus laterosporus Arthrobacter agilis Bacillus amyloliquefaciens Alkalihalophilus pseudofirmus Bacillus velezensis Bacillus subtilis subsp. natto Heyndrickxia oleronia Bacillus pumilus Shouchella clausii Cellulosimicrobium cellulans Bacillus licheniformis Cellulomonas fimi Lentibacillus amyloliquefaciens Clostridium beijerinckii Corynebacterium stationis Heyndrickxia coagulans Micrococcus luteus Clostridium butyricum Lactiplantibacillus plantarum Bifidobacterium longum subsp. infantis Bifidobacterium breve Bifidobacterium pseudocatenulatum Enterococcus faecalis Bifidobacterium longum subsp. longum Enterococcus faecium Lacticaseibacillus paracasei Lactococcus cremoris Bifidobacterium longum Lactiplantibacillus pentosus | Bifidobacterium breve Bifidobacterium pseudocatenulatum Bifidobacterium longum subsp. infantis Bifidobacterium bifidum Bifidobacterium longum Bifidobacterium catenulatum Bifidobacterium adolescentis Bifidobacterium longum subsp. longum Bifidobacterium animalis subsp. lactis Pediococcus pentosaceus Pediococcus acidilactici Lactobacillus acidophilus Brevibacillus brevis Escherichia coli Lactobacillus delbrueckii subsp. bulgaricus Limosilactobacillus reuteri Lactobacillus gasseri Lactobacillus jensenii Lactobacillus johnsonii Enterococcus durans Lactobacillus helveticus Pseudomonas putida Streptococcus thermophilus Limosilactobacillus fermentum Ligilactobacillus salivarius Levilactobacillus brevis Lactobacillus kefiranofaciens Lactobacillus crispatus Lentilactobacillus kefiri Leuconostoc mesenteroides Arthrobacter agilis Micrococcus luteus Lactococcus cremoris Leuconostoc lactis Alkalihalophilus pseudofirmus Lactococcus lactis Priestia megaterium Corynebacterium stationis Acinetobacter calcoaceticus Anaerobutyricum hallii Brevibacillus laterosporus Lactiplantibacillus plantarum Streptomyces fradiae Pimelobacter simplex Cellulomonas fimi Lactiplantibacillus pentosus Bacillus licheniformis Lacticaseibacillus casei Lacticaseibacillus rhamnosus Lentibacillus amyloliquefaciens |
Some probiotics are high on all three lists, for example: E.Coli. Others are not. I am inclined to using enzymes as the preferred abstraction. Metabolites have a very nice clustering, but at present deriving probiotics is not as clean and simple as desired. A more complex model is needed.
What have we learnt:
- There may not be studies on probiotics for a specific condition
- There are studies on probiotics that shifts some taxa. Things can become complex when there are multiple taxa in scope (as well as reliability of taxa identification)
- From the KEGG Enzymes estimated from a sample, we can derive the enzyme producing probiotics that may conceptually help
- Note: Organic Acid Test (OATS) report on many of these enzymes and can be used to validate estimates. Additionally, OATS tests can be used to select probiotics for the reported deficiencies
- From the KEGG metabolites estimated from a sample, we can supplement with the deficiency where practical, or look for probiotics that produces deficient metabolites.
The Enzymes and Metabolite approaches should produce reasonable candidates for future clinical studies.
Patient Specific Suggestions
The above exploration analysis was done ignoring the amount of bacteria in a specific example (and thus enzymes and metabolites). It also ignored whether there is duplication of enzymes and metabolites in the probiotics. Ideally, you want a full coverage of enzymes and metabolites.
Recent Comments