After the post on histamines, people with Salicylate sensitivity started to contact. “Me too!”. I looked at the data available and was disappointed that only 31 people have annotated their sample with this condition.
Medical Literature
This is known as salicylate allergy, salicylate hypersensitivity or salicylate intolerance. The traditional medical view is “It’s not clear exactly what causes salicylate allergy. “[WebMD] It was associated with Lyme Disease in 1991, and in other studies to: rheumatic diseases [1961] and systemic lupus erythematosus[1979]. It is seen in up to 7% of all patients with inflammatory bowel syndrome[2004] where as it was been estimated that 2.5%[2015] of the general population has this issue. The incidence in samples uploaded to microbiome prescription with symptom annotations appears to be 1.8%. This implies a significant microbiome contribution to this condition.
In 1980, we have The uselessness of blood salicylate levels in the diagnosis of salicylate hypersensitivity. However, the level may cause issues with blood donations being received by people with this condition, see Salicylate and acetaminophen in donated blood [1986].
Diet changes appear to have minor impact as reported in Effectiveness of Personalized Low Salicylate Diet in the Management of Salicylates Hypersensitive Patients: Interventional Study[2021] and others [2021] [2016]
In dealing with Plants we have an interesting study Overexpression of Arabidopsis NIMIN1 results in salicylate intolerance [2016] with other plant based studies [2010].
Human Trials
Only one trial could be found
After dietary supplementation with 10 g daily of fish oils rich in omega-3 PUFAs for 6-8 weeks all three experienced complete or virtually complete resolution of symptoms allowing discontinuation of systemic corticosteroid therapy. Symptoms relapsed after dose reduction.”
Control of salicylate intolerance with fish oils [2008]
KEGG: Kyoto Encyclopedia of Genes and Genomes Perspective
I am by temperament, a modeler. Look at the surface layer of information, derive some suggestions of how thing work and then, critically, test the model. Failure is information also! My first model is to assume enzymes associated with salicylates are significant.
| Salicylate aka o-Hydroxybenzoic acid aka Salicylic acid is compound C00805 with the following enzymes interacting with it. Conceptually, too much of some enzymes and too few of other enzymes may be the likely cause. The list include the followin: |
- 1.2.1.65 salicylaldehyde dehydrogenase
- 1.14.13.1 salicylate 1-monooxygenase;
- 1.14.13.172 salicylate 5-hydroxylase
- 2.1.1.274 salicylate 1-O-methyltransferase
- 3.1.1.55 acetylsalicylate deacetylase
- 4.1.1.91 salicylate decarboxylase
- 4.2.99.21 isochorismate lyase
- 6.2.1.61 salicylate—[aryl-carrier protein] ligase
- 6.2.1.65 salicylate—CoA ligase
Data From Microbiome Prescription Samples
See Special Studies on Symptoms caused by Bacteria – v.2 for technical detail. A related post is Histamine Release – Literature Review And Speculation.
For more information on each enzyme see BRENDA or KEGG
The most significant ones from are shown below. None are a match for the candidate list above
| EC | Enzyme Name | With Sal | Without Sal | TScore | DF | Probability |
| 1.1.1.337 | (2S)-2-hydroxycarboxylate:NAD+ oxidoreductase | 37077 | 19517 | 3.38 | 666 | P < 0.001 |
| 1.3.7.1 | 6-oxo-1,4,5,6-tetrahydronicotinate:ferredoxin oxidoreductase | 334 | 91 | 4.01 | 222 | P < 0.001 |
| 2.1.1.245 | 5-methyltetrahydrosarcinapterin:corrinoid/iron-sulfur protein methyltransferase | 36498 | 18608 | 3.46 | 666 | P < 0.001 |
| 2.1.1.258 | 5-methyltetrahydrofolate:corrinoid/iron-sulfur protein methyltransferase | 36569 | 18689 | 3.46 | 666 | P < 0.001 |
| 2.6.1.109 | 8-amino-3,8-dideoxy-alpha-D-manno-octulosonate:2-oxoglutarate aminotransferase | 108 | 35 | 3.90 | 124 | P < 0.001 |
| 2.7.7.39 | CTP:sn-glycerol-3-phosphate cytidylyltransferase | 60861 | 32715 | 3.56 | 666 | P < 0.001 |
| 3.4.19.11 | gamma-D-glutamyl-meso-diaminopimelate peptidase | 39254 | 21501 | 3.42 | 666 | P < 0.001 |
| 3.5.2.18 | 6-oxo-1,4,5,6-tetrahydronicotinate amidohydrolase | 334 | 93 | 3.93 | 215 | P < 0.001 |
| 4.2.1.43 | 2-dehydro-3-deoxy-L-arabinonate hydro-lyase (2,5-dioxopentanoate-forming) | 433 | 103 | 4.57 | 214 | P < 0.001 |
| 5.4.99.21 | 23S rRNA-uridine2604 uracil mutase | 63603 | 39414 | 3.33 | 666 | P < 0.001 |
Looking at the next level of association, we still find none of these candidate enzymes.
| EC | Enzyme Name | With Sal | Without Sal | TScore | DF | Probability |
| 1.1.1.135 | GDP-6-deoxy-alpha-D-talose:NAD(P)+ 4-oxidoreductase | 279 | 96 | 2.82 | 263 | P < 0.01 |
| 1.1.1.312 | 4-carboxy-2-hydroxymuconate semialdehyde hemiacetal:NADP+ 2-oxidoreductase | 353 | 105 | 2.68 | 229 | P < 0.01 |
| 1.1.1.410 | D-erythronate:NAD+ 2-oxidoreductase | 381 | 117 | 3.32 | 300 | P < 0.01 |
| 1.1.99.31 | (S)-mandelate:acceptor 2-oxidoreductase | 265 | 92 | 2.71 | 210 | P < 0.01 |
| 1.13.11.37 | hydroxyquinol:oxygen 1,2-oxidoreductase (ring-opening) | 268 | 88 | 2.66 | 247 | P < 0.01 |
| 1.17.1.4 | xanthine:NAD+ oxidoreductase | 34795 | 17231 | 2.81 | 666 | P < 0.01 |
| 1.17.9.1 | 4-methylphenol:oxidized azurin oxidoreductase (methyl-hydroxylating) | 198 | 83 | 3.00 | 338 | P < 0.01 |
| 1.2.1.87 | propanal:NAD+ oxidoreductase (CoA-propanoylating) | 25104 | 13840 | 2.72 | 666 | P < 0.01 |
| 1.2.7.4 | carbon-monoxide,water:ferredoxin oxidoreductase | 51730 | 29588 | 3.18 | 666 | P < 0.01 |
| 1.2.99.7 | aldehyde:acceptor oxidoreductase (FAD-independent) | 34901 | 17791 | 2.71 | 666 | P < 0.01 |
| 1.3.1.32 | 3-oxoadipate:NAD(P)+ oxidoreductase | 268 | 88 | 2.65 | 260 | P < 0.01 |
| 2.3.1.101 | formylmethanofuran:5,6,7,8-tetrahydromethanopterin 5-formyltransferase | 289 | 88 | 3.25 | 207 | P < 0.01 |
| 2.3.1.276 | acetyl-CoA:alpha-D-galactosamine-1-phosphate N-acetyltransferase | 51215 | 29509 | 3.08 | 666 | P < 0.01 |
| 2.3.2.13 | protein-glutamine:amine gamma-glutamyltransferase | 49247 | 27040 | 3.22 | 666 | P < 0.01 |
| 2.4.1.245 | NDP-alpha-D-glucose:D-glucose 1-alpha-D-glucosyltransferase | 111 | 42 | 3.33 | 213 | P < 0.01 |
| 2.7.1.215 | ATP:erythritol 1-phosphotransferase | 12084 | 5590 | 2.73 | 653 | P < 0.01 |
| 2.7.4.31 | ATP:[5-(aminomethyl)furan-3-yl]methyl-phosphate phosphotransferase | 289 | 88 | 3.25 | 207 | P < 0.01 |
| 2.7.4.33 | ADP:polyphosphate phosphotransferase | 36369 | 19252 | 2.68 | 666 | P < 0.01 |
| 2.7.7.83 | UTP:N-acetyl-alpha-D-galactosamine-1-phosphate uridylyltransferase | 51217 | 29512 | 3.08 | 666 | P < 0.01 |
| 2.7.9.6 | ATP:rifampicin, water 21-O-phosphotransferase | 1458 | 427 | 2.99 | 575 | P < 0.01 |
| 3.1.1.104 | 5-phospho-D-xylono-1,4-lactone hydrolase | 12119 | 5680 | 2.70 | 656 | P < 0.01 |
| 3.1.1.81 | N-acyl-L-homoserine-lactone lactonohydrolase | 12121 | 5591 | 2.73 | 653 | P < 0.01 |
| 3.4.23.43 | prepilin peptidase | 104657 | 72966 | 2.58 | 666 | P < 0.01 |
| 3.5.1.110 | (Z)-3-ureidoacrylate amidohydrolase | 12177 | 5917 | 2.63 | 661 | P < 0.01 |
| 3.5.4.44 | ectoine aminohydrolase | 268 | 85 | 2.71 | 234 | P < 0.01 |
| 3.5.99.5 | 2-aminomuconate aminohydrolase | 309 | 106 | 2.72 | 177 | P < 0.01 |
| 3.6.1.15 | nucleoside-triphosphate phosphohydrolase | 26554 | 15567 | 3.28 | 665 | P < 0.01 |
| 3.6.1.7 | acylphosphate phosphohydrolase | 60707 | 38253 | 2.67 | 666 | P < 0.01 |
| 4.2.1.147 | 5,6,7,8-tetrahydromethanopterin hydro-lyase (formaldehyde-adding, tetrahydromethanopterin-forming) | 309 | 101 | 2.77 | 192 | P < 0.01 |
| 5.3.1.15 | D-lyxose aldose-ketose-isomerase | 58198 | 37194 | 2.64 | 666 | P < 0.01 |
| 5.3.1.34 | D-erythrulose-4-phosphate ketose-aldose isomerase | 12039 | 5560 | 2.71 | 642 | P < 0.01 |
| 7.5.2.4 | ATP phosphohydrolase (ABC-type, teichoic-acid-exporting) | 25268 | 12222 | 2.91 | 666 | P < 0.01 |
So, model 1 is a bust — none of the suspected enzymes showed up as being statistically significant. This echoes The uselessness of blood salicylate levels in the diagnosis of salicylate hypersensitivity[1980]. However one thing stands out…. for each of the enzymes estimates above — the amount with salicylate sensitivity is in every case more than those without it. This is unusual.It hints that the issue is over production of enzymes.
Microbiome Significances
Only Biomesight has sufficient data to do an investigation with. The most significant ones all had an interesting characteristic matching that seen with the enzymes — Too MANY.
| Tax_name | Tax_rank | With Sal | Without Sal | Tscore | DF | Probability |
| Fusobacteriia | class | 21983 | 3330 | 3.52 | 346 | P < 0.001 |
| Chroococcaceae | family | 588 | 165 | 3.42 | 286 | P < 0.001 |
| Fusobacteriaceae | family | 25628 | 3687 | 3.63 | 305 | P < 0.001 |
| Hungateiclostridiaceae | family | 8534 | 2649 | 3.69 | 201 | P < 0.001 |
| Lachnospiraceae | family | 297116 | 203706 | 4.2 | 667 | P < 0.001 |
| Rhodanobacteraceae | family | 2453 | 656 | 3.32 | 478 | P < 0.001 |
| Syntrophobacteraceae | family | 115 | 36 | 3.57 | 184 | P < 0.001 |
| Acetobacterium | genus | 3977 | 1250 | 3.92 | 657 | P < 0.001 |
| Blautia | genus | 141777 | 87110 | 4.2 | 667 | P < 0.001 |
| Chroococcus | genus | 588 | 165 | 3.42 | 286 | P < 0.001 |
| Clostridium | genus | 30531 | 18614 | 3.45 | 667 | P < 0.001 |
| Furfurilactobacillus | genus | 91 | 38 | 3.48 | 270 | P < 0.001 |
| Hungateiclostridium | genus | 8178 | 2229 | 3.94 | 200 | P < 0.001 |
| Luteibacter | genus | 3734 | 764 | 4.07 | 387 | P < 0.001 |
| Fusobacteriales | order | 21983 | 3329 | 3.52 | 346 | P < 0.001 |
| Syntrophobacterales | order | 135 | 45 | 5.06 | 331 | P < 0.001 |
| Fusobacteria | phylum | 21983 | 3330 | 3.52 | 346 | P < 0.001 |
| Bacteroides fluxus | species | 2722 | 318 | 3.41 | 517 | P < 0.001 |
| Blautia glucerasea | species | 4228 | 664 | 3.86 | 550 | P < 0.001 |
| Blautia schinkii | species | 1710 | 265 | 4.14 | 558 | P < 0.001 |
| Caloramator uzoniensis | species | 300 | 85 | 4.53 | 328 | P < 0.001 |
| Chroococcus minutus | species | 588 | 165 | 3.42 | 286 | P < 0.001 |
| Clostridium akagii | species | 113 | 43 | 4.37 | 311 | P < 0.001 |
| Furfurilactobacillus siliginis | species | 91 | 38 | 3.51 | 271 | P < 0.001 |
| Luteibacter anthropi | species | 3734 | 764 | 4.07 | 387 | P < 0.001 |
Looking at the next level, we have a lot less BUT THE PATTERN OF OVER ABUNDANCE continues:
| Tax_name | Tax_rank | WithMean | WithoutMean | Tscore | DF | Probability |
| Desulfovibrionaceae | family | 8811 | 5037 | 2.61 | 641 | P < 0.01 |
| Xanthomonadaceae | family | 2131 | 642 | 3.16 | 556 | P < 0.01 |
| Bilophila | genus | 6608 | 3415 | 3.25 | 587 | P < 0.01 |
| Caldicellulosiruptor | genus | 581 | 289 | 2.67 | 617 | P < 0.01 |
| Fusobacterium | genus | 2795 | 683 | 2.76 | 289 | P < 0.01 |
| Holdemania | genus | 572 | 296 | 3.04 | 589 | P < 0.01 |
| Lachnospira | genus | 49628 | 28041 | 3.22 | 667 | P < 0.01 |
| Macrococcus | genus | 2425 | 419 | 3.28 | 222 | P < 0.01 |
| Thiorhodococcus | genus | 64 | 28 | 2.67 | 108 | P < 0.01 |
| Desulfovibrionales | order | 8840 | 5039 | 2.63 | 643 | P < 0.01 |
| Bacteroides finegoldii | species | 7458 | 2163 | 3.15 | 552 | P < 0.01 |
| Blautia obeum | species | 12039 | 5596 | 2.68 | 642 | P < 0.01 |
| Fusobacterium gonidiaformans | species | 3980 | 833 | 3.12 | 134 | P < 0.01 |
| Shewanella upenei | species | 92 | 46 | 3 | 157 | P < 0.01 |
Cross Validation of The above
We know of only one thing known to help, Omega-3 / Fish Oil. We have this information in our database and find that it decreases the following (from multiple studies):
- Lachnospira
- Lachnospiraceae
- Furfurilactobacillus siliginis
- Blautia
A single study suggests that it increases Clostridium / Clostridium akagii (Child). This is a 80% validation rate implying that the Artificial Intelligence suggestions on a person’s microbiome is likely to be effective.
Putting this data to Use
The first step is simple, take 10 g daily of fish oils rich in omega-3 for the first 6-8 weeks. Get relief. Warning: the study states that if you stop, you will revert. It is insufficient to correct the bacteria shifts.
If you do not have a BiomeSight sample on hand…
Go to Citizen Science Special Studies, Enter Salicylate in the search box. The page should give you two buttons as shown below.
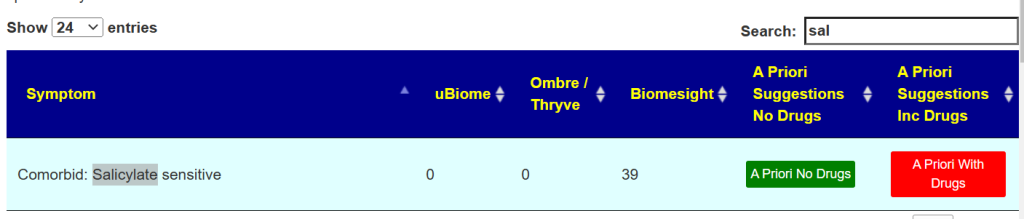
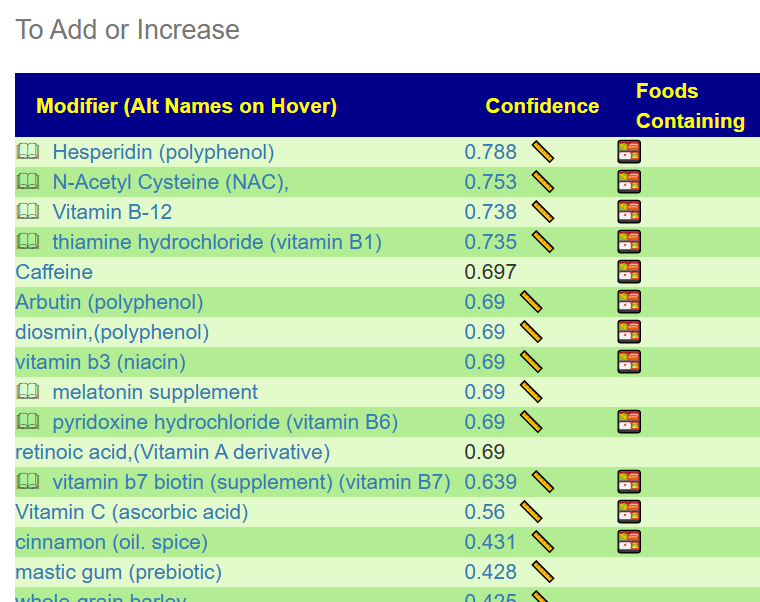
The results may change as more samples are uploaded and annotated with symptoms. This research is dynamic and live!. Clicking the green button will show a list of item to take or add to your diet.
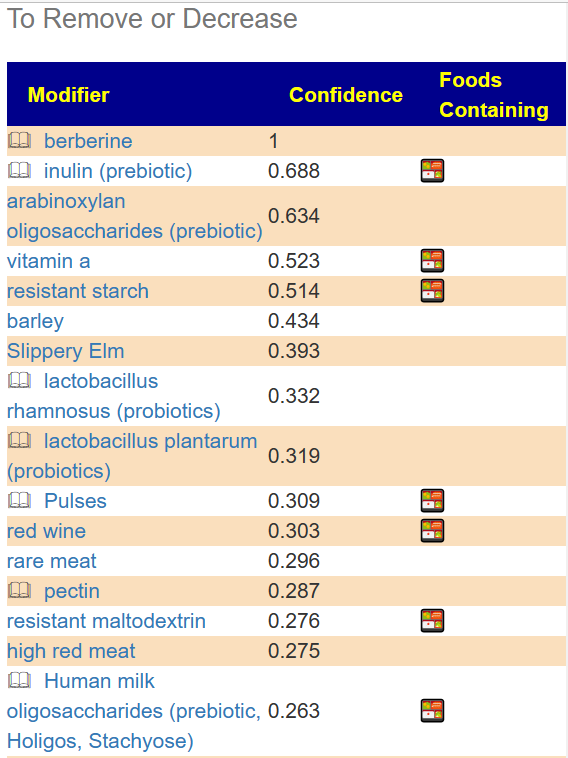
The second item is likely more important items that feed the bacteria that are too high. You want to reduce or eliminate these.
There is still more information available on actual diet (foods that you may eat), by clicking on

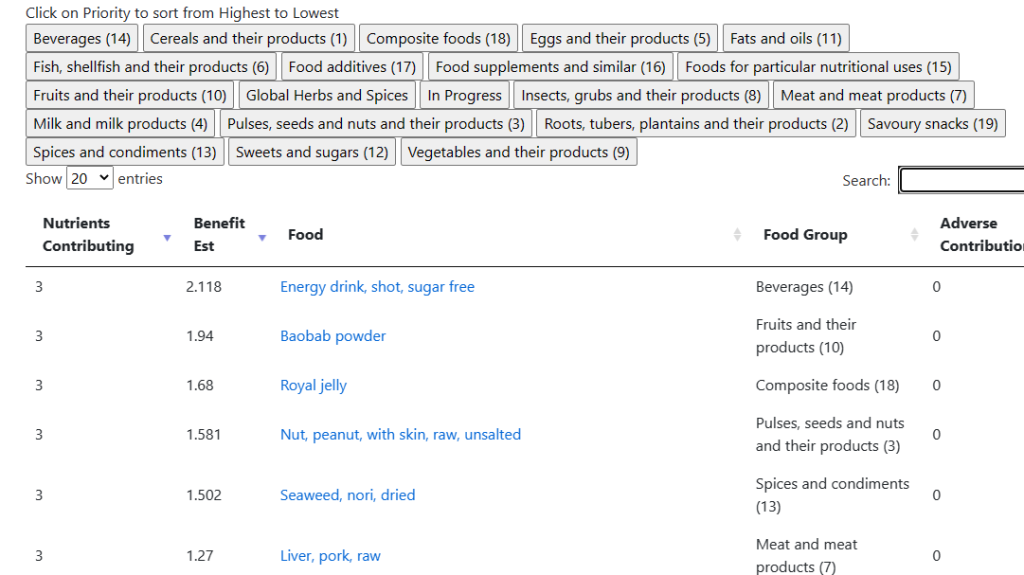
We know the fish oil helps, let us verify that by clicking on [FIsh, shellfish and their products]. We see fish — specifically salmon! And Walrus liver is a to-avoid!

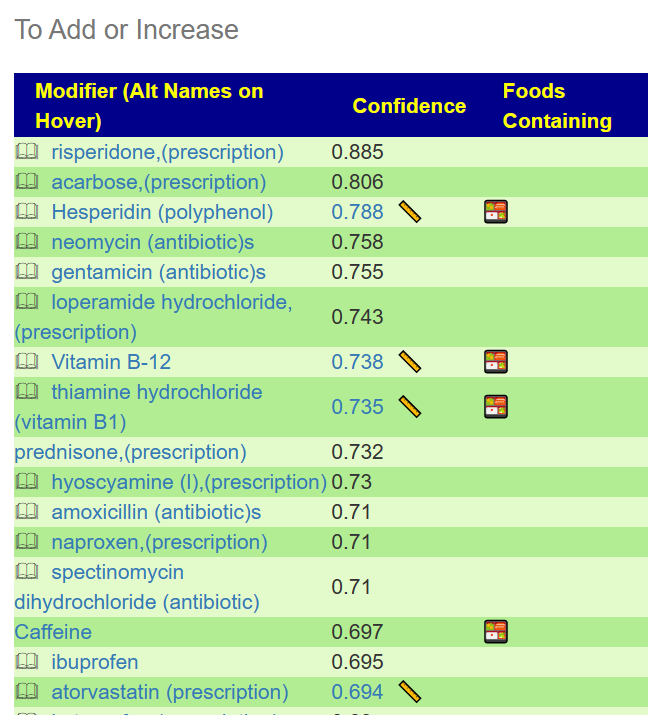
Returning to the red button, we see a list of off-label usage of drugs mixed into the results. The top ones:
- Risperidone is used to treat a certain mental/mood disorder called schizophrenia.
- Acarbose is used with a proper diet and exercise program to control high blood sugar in people with type 2 diabetes.
- meomycin is used to decrease the risk of infection after certain intestinal surgeries.
Step 2 Order a Suitable Test
You will likely need at least 3 tests, the data is based on Biomesight tests. Order some. If you live in the US, you could order an Ombre Test (this gets more technical because you will need to download some big files [FASTQ] from their site and then upload to Biomesight for them to process them).
You will likely be doing the above for 5-6 weeks before you get your data available on BiomeSight.
Log in to Microbiome Prescription
Go to [Research Features]
Then find on that page, the Experimental section, and click v.2 Of Special Symptom Associations

Select your latest sample (if more than one) and then the condition

The bacteria that definitely match the pattern will be automatically checked.
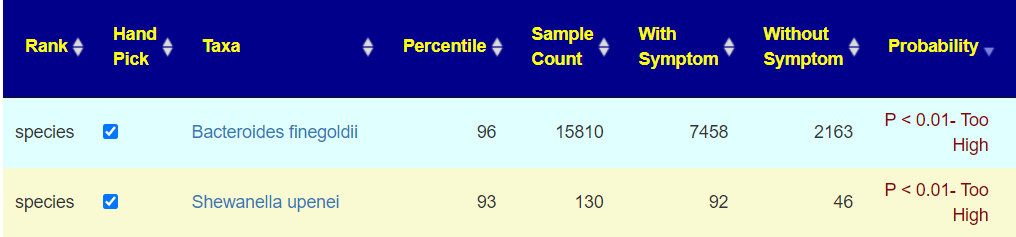
You should scan for any close matches and check those boxes. For example, above the without symptoms slightly.
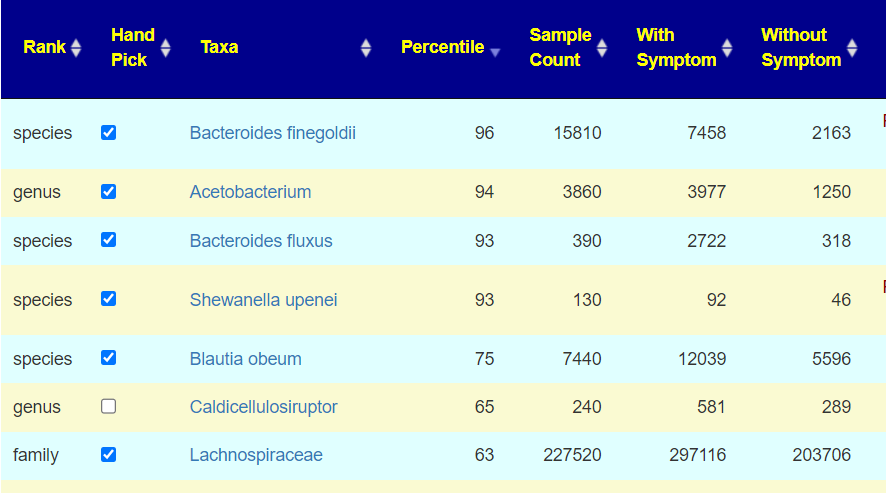
Now click the [Add to Hand Picked LIst] button at the top
A new window will appear with the items that you have selected.
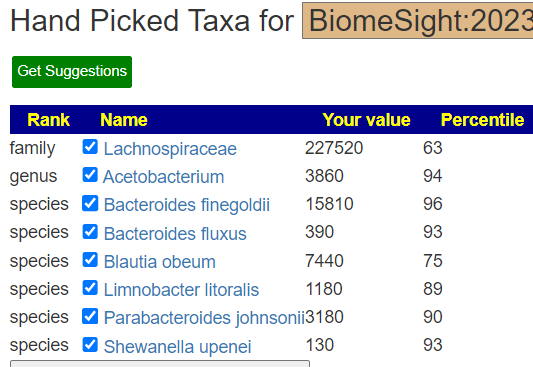
Just click [Get Suggestions] and then scope the type of suggestions you wish
Then click the Get Suggestions button on the top left
Your suggestions may be similar (the sample that I am using was for someone with this issue)
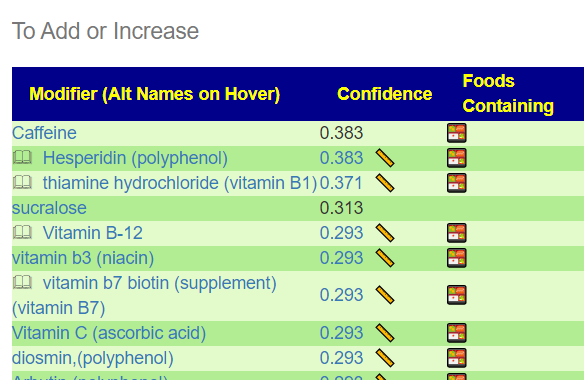
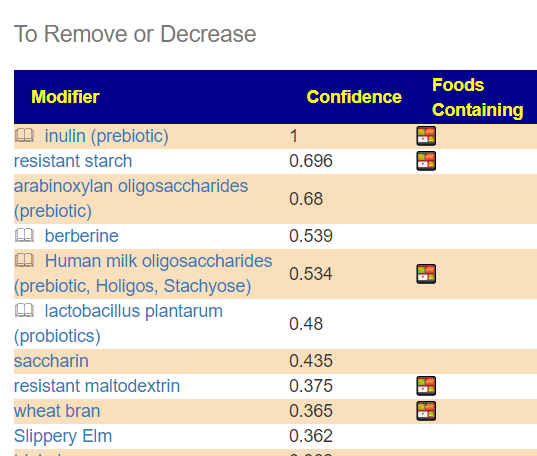
The food list may also change
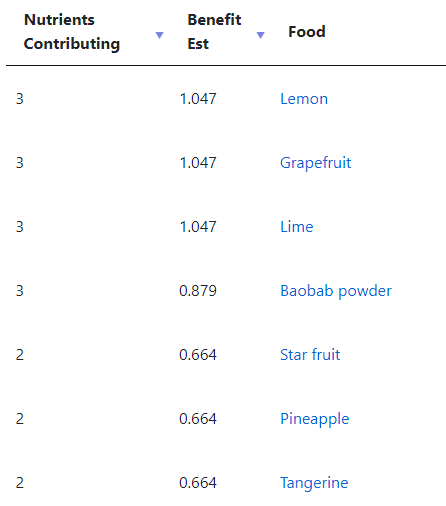
WARNING: Many items suggested may trigger a Salicylate reaction. Consider doing the 6-8 weeks of Fish Oil before starting any items that could trigger a reaction.
This reaction could be caused by bacteria resisting change (similar to a Jarisch Herxheimer)
The usual pattern is to implement the changes for 4 week, do a new sample. Keep doing the changes until the results come in. So this is the likely time line for some
- Week 0: Order Kit, start general list
- Week 1: Take test #1 and send in
- Week 4: Get results, upload and get 1st personalized set of suggestions
- Week 8: Take test #2 and send in
- Week 10: Get results, upload and get 2nd personalized set of suggestions
- Week 14: Take test #3 and send in
- Week 16: Get results, upload and get 3rd personalized set of suggestions
Depending on results, repeat. Your desired result is to have NO Matches with the pattern (and hopefully have eliminated the issue).
Postscript – and Reminder
I am not a licensed medical professional and there are strict laws where I live about “appearing to practice medicine”. I am safe when it is “academic models” and I keep to the language of science, especially statistics. I am not safe when the explanations have possible overtones of advising a patient instead of presenting data to be evaluated by a medical professional before implementing.
I cannot tell people what they should take or not take. I can inform people items that have better odds of improving their microbiome as a results on numeric calculations. I am a trained experienced statistician with appropriate degrees and professional memberships. All suggestions should be reviewed by your medical professional before starting.
The answers above describe my logic and thinking and is not intended to give advice to this person or any one. Always review with your knowledgeable medical professional.
1 thought on “Salicylate sensitive – Data And Research”
Comments are closed.