Histamine issues can occur from consuming food high in histamines. This is the typical approach for people dealing with this issue. There is an another route that should not be overlooked — things that do not contain histamine but which triggers histamine release.
Citations
Histamine release caused by reactions to drug product and/or excipients/vehicles is a phenomenon observed in both toxicology and pharmacology studies. This type of reaction is also referred to as pseudoallergic reaction, anaphylactoid reaction or complement activation-related pseudoallergy (CARPA).
Biomarkers in Toxicology, 2014
- “Codeine and meperidine are examples of other opioids that can induce mast cell activation with the release of histamine” [ 2020]
- “Quaternary ammonium compounds (e.g., NMBDs) are generally weaker histamine-releasing substances than are tertiary amines such as morphine.”
- “Histamine release can occur with administration of certain opioids” [2015]
- “Histamine release is primarily caused by morphine, followed by hydromorphone,” [2009]
- “Histamine release and the severity of reactions during vancomycin administration are directly dependent on the rate of infusion.” [2007]
- “It is not clear whether histamine levels are altered following hypo– or hyperthermia seen during several clinical or experimental situations.” [2004]
- “Histamine release and non-IgE-mediated anaphylactic (anaphylactoid) reactions occur with alcuronium” [2016]
The above means that care needs to be taken if a herb, spice or supplement causes a histamine reaction. It may not be histamine in the substance, rather the substance causes mast cells to react and dump histamine. It is a significantly different situation. Other items that have been reported to cause histamine release include: Vitamin C(L-Ascorbic acid), Niacin (vitamin B3), Quercetin, Stinging nettle, Licorice root, Ginkgo biloba, Chamomile and Echinacea.
Histamine toxicity is sometimes confused with an allergic reaction to fish. Here is why:
American Academy of Allergy, Asthma & Immunology
Some kinds of fish contain naturally high levels of the chemical histidine. This chemical can be converted to histamine by bacteria [ the enzyme histidine decarboxylase EC 4.1.1.22]. In an allergic reaction, mast cells release histamine which triggers allergy symptoms. So, if a person eats fish that has a high level of histamine, the response may resemble an allergic reaction to that food.
A list of bacteria with this enzyme is here. The data comes from the KEGG: Kyoto Encyclopedia of Genes and Genomes and a collection of several thousand samples from different labs uploaded to the Microbiome Prescription web site. The top producers over all of the sample are shown below
| Bacteria | Estimate Occurances | Contribution |
|---|---|---|
| Bacteroides fragilis [species] | 1 | 5897.04 |
| Eggerthella lenta [species] | 1 | 627.29 |
| Acinetobacter baumannii [species] | 0.9 | 109.55 |
| Gordonibacter pamelaeae [species] | 1 | 99.28 |
| Fusobacterium varium [species] | 1 | 93.35 |
| Clostridium perfringens [species] | 1 | 64.31 |
| Fusobacterium ulcerans [species] | 1 | 57.3 |
Hypothesis Testing on Bacteria Conversion
The citizen science site, Microbiome Prescription, allows people to share their microbiome results from many labs and to annotate their samples with symptoms. Over 1000 samples have these annotations as shown below, so we can suggest a hypothesis and test it.
Hypothesis: People with Histamine Issues are likely to have higher counts of bacteria producing histidine decarboxylase
The results was a bit of a surprise. The Hypothesis failed dramatically! Having more bacteria producing this enzyme appears to be associated with less histamine issue!! This pattern persists across all three labs with significant data sample size.
One hypothesis that could be suggested by this data is that the histamine issue is due to the body’s base level of histamine being abnormally low and thus the body is unfamiliar with histamines and thus overreacts.
An Analogy to Consider
Imagine someone whose diet lacks ANY added sugar. After a year, he drops into a friend who makes him his favorite herbal tea. The friend, she, likes sweet tea and adds several teaspoon of sugar. This person drinks it and gets an atypical headache which confuses the friend – he drinks it regularly! The real cause is too low a base level of sugar consumption for this person to tolerate.
The above suggests that the same may be occurring with histamine reactions.
Did you know that both too much sugar and too little of sugar can cause headaches? When you consume too much sugar at once or don’t eat for an extended period of time, you can cause rapid fluctuations in your blood sugar levels which can trigger a headache. Some people are more prone to these sugar-triggered headaches.
Source
If this is a correct speculation, then the treatment approach is to encourage bacteria that produce the enzyme histidine decarboxylase EC 4.1.1.22.
Phrase 2 Checking each bacteria Taxa
While a bacteria has an enzyme, it is not a given that it is active. Our second pass is looking at the frequency of each bacteria taxa appearing in each group. If a specific taxa is significantly more or less frequent, it may be a trail worth following. Drilling down to species causes our observations to drop to the point that many data points cannot be examined for significance. We have three taxa with good sample sizes.
Remember, different labs use different software resulting in different taxa names.
| Bacteria | Percent Having | Mean | SD | Subset | Lab |
| Bacteroides fragilis | 79.2 | 5138 | 8629 | Histamine | BiomeSight |
| 81.4 | 6369 | 16168 | No Histamine | ||
| 75.6 | 6736 | 22788 | Histamine | uBiome | |
| 51.8 | 16628 | 47841 | No Histamine | ||
| 94.2 | 7343 | 14279 | Histamine | Ombre | |
| 94.2 | 8646 | 27095 | No Histamine | ||
| Eggerthella lenta | 74 | 57 | 611 | Histamine | BiomeSight |
| 69.8 | 365 | 346 | No Histamine | ||
| 74.4 | 67 | 1323 | Histamine | uBiome | |
| 69.5 | 235 | 1502 | No Histamine | ||
| 77.9 | 824 | 2407 | Histamine | Ombre | |
| 79.4 | 1059 | 4131 | No Histamine | ||
| Gordonibacter pamelaeae | 57.78 | 52 | 459 | Histamine | uBiome |
| 61.2 | 207 | 413 | No Histamine | ||
| 51.9 | 124 | 177 | Histamine | Ombre | |
| 46.1 | 187 | 358 | No Histamine |
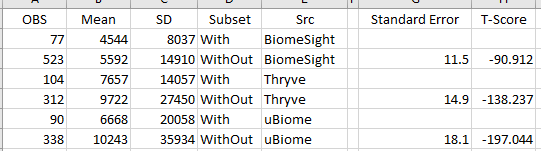
Bacteroides fragilis for Ombre(Thryve), uBiome and BiomeSight were the most statistically significant and all have the same pattern: People reporting histamine issues had less than people that did not report histamine issues. Bacteroides fragilis is also the main source of histidine decarboxylase from the microbiome.
uBiome data was very interesting because the detection rate for Bacteroides fragilis was significantly less for samples that did not report histamine issues with the differences of means being much much more than with other labs. This hints that some part of the base pairs collection that uBiome used to determine Bacteroides fragilis in their software may be particularly important for histamine issues. To put it another way, some part of the sequence being used to determine the taxa, also appears to detect histamine issues. This leads to the possibility that specific strains of Bacteroides fragilis may result in better histamine tolerance.
uBiome and others use a reference database. Because a 16s test only looks at a tiny portion of the microbial genome, of necessity different bioinformatics pipelines will assign slightly different microbial genus/species names for the string of base pairs they sequence.
uBiome, a company that offered microbiome testing services, used a proprietary reference database called the uBiome Microbial Insights Test (MIT) reference database for the taxonomic classification of 16S rRNA gene sequences. This database was specifically designed for the analysis of human microbiome samples, and included over 1,000 microbial taxa that were commonly found in the human gut, oral, and skin microbiomes. The database was built using a combination of publicly available 16S rRNA gene sequences and uBiome’s own sequencing data, and was regularly updated to incorporate new microbial taxa and improve the accuracy of taxonomic assignments.
From Chat_GPT
- Biomesight: SILVA SSU Ref NR 99 database,
- Ombre: GreenGenes 13_8 release
- American Gut: Greengenes 13_8 release,
- Thorne: Their 16s database is built from a combination of publicly available reference databases, including the Greengenes and SILVA databases, as well as Thorne’s own sequencing data.
Unfortunately uBiome reference database disappeared with it going into bankruptcy. Only time will tell if Thorne’s reference database will identify the key base pairs that appears to be connected to histamine issues.
What is very interesting comes from a 1999 study, “Intestinal mucosa-associated bacteria modulate rat mast cell reactivity” which reports a ConA-induced histamine release was diminished up to 71% of maximal histamine release by Bacteroides fragilis!! This supports that increasing Bacteroides fragilis may be the right way to go.
Where do we go from here for bacteria?
The above suggests that there is a bacteria taxa with a specific key base pair that is connected to histamines issues. Let us call it “Bacteroides Histamilis”(BH). This taxa has some overlap with Bacteroides fragilis (BF). They appear to occupy a similar niche in the microbiome world with an increase of BF decreasing BH (or it’s impact). We are shooting in the fog here, but it seems encouraging BF growth may reduce BH and ease histamine issues. We have know items that increases or decrease listed here: Bacteroides fragilis and a link to possible foods. The top food nutrients are iron, zinc and Riboflavin (Vitamin B2).
Going to Chat_GPT for some quick answers, we get a pleasant surprise:
Iron plays an important role in the regulation of histamine levels in the body. It is required for the activity of an enzyme called diamine oxidase (DAO), which is responsible for breaking down histamine in the gut. Low iron levels can lead to reduced DAO activity and increased histamine levels, which can contribute to histamine intolerance or sensitivity.
Zinc is also important for the activity of DAO, as well as for the regulation of histamine receptors in the body. Zinc deficiency has been linked to increased histamine levels and may contribute to histamine intolerance.
Riboflavin (vitamin B2) is required for the synthesis of DAO, and low riboflavin levels have been associated with reduced DAO activity and increased histamine levels.
Answer from Chat_CPT to question: “Is histamine sensitivity connected to iron, zinc or Riboflavin deficiency?“
So, we have an interesting cascade… the 3 key nutrients available concurrently in food that increases Bacteroides fragilis, are all associated with DAO production. Should we speculate that BH is a mutation that thrives better with low levels of Iron, Zinc and Riboflavin and the difference in base pairs is connected with this mutation? Continuing this thought experiment, would this mutation also have reduced (or no) enzyme histidine decarboxylase EC 4.1.1.22 being produced resulting in an abnormally low level of histamine on an ongoing basis and thus increased sensitivity?
A Parallel Thread in Autism?
Antihistamines have been reported to reduce some autism behaviors [2018]. For example “Altered expression of histamine signaling genes in autism spectrum disorder” [2017]. Bacteroides fragilis has been reported to be low with autism. I will leave it to others to explore this further.
One study published in 2013 found that children with autism had lower levels of Bacteroides fragilis in their gut microbiome compared to typically developing children. Another study published in 2017 found that a group of children with autism had higher levels of Bacteroides fragilis in their fecal samples compared to a control group of typically developing children.
Iron plays an important role in brain development and function, and some studies have suggested that iron deficiency during pregnancy or early childhood may increase the risk of autism.
Zinc is also important for brain development and function, and some studies have found that children with autism may have lower levels of zinc in their blood or hair compared to typically developing children.
Riboflavin (vitamin B2) is required for several important metabolic pathways in the body, and some studies have suggested that children with autism may have lower levels of riboflavin compared to typically developing children.
However, other studies have not found a significant association between these and autism.
Chat_GPT.
Bottom Line
There is a scent that specific strains of Bacteroides fragilis may be associated with histamine sensitivity. In an environment deficient of iron, zinc or riboflavin, this strain increases. This strain may not produce histidine decarboxylase EC 4.1.1.22 (epigenetics?) resulting in a much lower level of histamine in the body resulting in “sugar-shock” when a food containing histamines is consumed. We saw above a consistent pattern of having a lower count has an increased probability of histamine issues. A lower count is typically viewed as having less appropriate nutrients available – and the missing nutrients are implied by our analysis.
A common pattern seen by people with Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS), Irritable Bowel Disease and other gut disturbances is increasing histamine issues. Mal-absorption due to gut disturbances would result in a dropping of iron, zinc and riboflavin absorption causing this “Bacteroides Histamilis” strain to dominant.
With this model, supplementation with iron, zinc and riboflavin to increase the body’s level to at least the 75%ile may result in significant improvement.
A suggestion for a study would be to measure iron, zinc and riboflavin levels of people with histamine issues against an appropriate matched control population. This may determine if deficiency of just one of this trio is sufficient, or do we need multiple deficiency.
Appendix Statistical Significance Table
The following is general data mining. Remember the lab’s software determines the taxa names using probability. Safest conclusions are when multiple labs report significance in the same direction for the same taxa. Taxa cannot be safely be applied to different labs. Labs report on different bacteria, especially at the species level, on occasion they will pick one name and a different lab will pick a different name.
REMEMBER: These may not be the cause, rather bacteria altered by the bacteria that are the cause.
| Taxa name | Taxa Rank | Lab | Histamine Mean | Control Mean | T-Score | DF | Probability |
| Cyanobacteria /Melainabacteria group | clade | uBiome | 6444 | 268 | 4.338711 | 12 | P < 0.001 |
| Alphaproteobacteria | class | uBiome | 26227 | 15210 | 3.283924 | 269 | P < 0.01 |
| Chitinophagia | class | Thryve | 94 | 69 | 2.824838 | 330 | P < 0.01 |
| Clostridia | class | BiomeSight | 619159 | 558435 | 2.713198 | 667 | P < 0.01 |
| Deltaproteobacteria | class | uBiome | 10291 | 6987 | 2.794664 | 401 | P < 0.01 |
| Flavobacteriia | class | Thryve | 377 | 93 | 4.138491 | 241 | P < 0.001 |
| Spirochaetia | class | Thryve | 22731 | 196 | 2.747053 | 68 | P < 0.01 |
| Bifidobacteriaceae | family | uBiome | 30890 | 14422 | 2.643228 | 474 | P < 0.01 |
| Carnobacteriaceae | family | uBiome | 815 | 138 | 2.86021 | 182 | P < 0.01 |
| Chitinophagaceae | family | Thryve | 94 | 69 | 2.818824 | 330 | P < 0.01 |
| Chromatiaceae | family | BiomeSight | 110 | 74 | 2.962499 | 525 | P < 0.01 |
| Coprobacillaceae Verbarg et al. 2014 | family | BiomeSight | 4986 | 2981 | 2.905239 | 662 | P < 0.01 |
| Desulfovibrionaceae | family | uBiome | 10288 | 6984 | 2.797459 | 401 | P < 0.01 |
| Flavobacteriaceae | family | Thryve | 376 | 95 | 3.766135 | 215 | P < 0.001 |
| Lachnospiraceae | family | BiomeSight | 244569 | 201657 | 3.461006 | 667 | P < 0.001 |
| Micrococcaceae | family | Thryve | 2041 | 279 | 2.732836 | 198 | P < 0.01 |
| Odoribacteraceae | family | uBiome | 13802 | 9604 | 2.953385 | 386 | P < 0.01 |
| Pasteurellaceae | family | uBiome | 11259 | 3343 | 2.952883 | 325 | P < 0.01 |
| Rhodospirillaceae | family | uBiome | 29014 | 17878 | 3.045209 | 229 | P < 0.01 |
| Rubritaleaceae | family | BiomeSight | 118 | 51 | 3.018423 | 231 | P < 0.01 |
| Streptococcaceae | family | uBiome | 12988 | 6609 | 3.294864 | 469 | P < 0.01 |
| Syntrophaceae | family | BiomeSight | 88 | 34 | 3.602406 | 95 | P < 0.001 |
| Weeksellaceae | family | Thryve | 826 | 140 | 2.707921 | 42 | P < 0.01 |
| Adlercreutzia | genus | BiomeSight | 856 | 346 | 3.933686 | 493 | P < 0.001 |
| Anaerofustis | genus | BiomeSight | 85 | 45 | 2.957077 | 132 | P < 0.01 |
| Anaerolinea | genus | BiomeSight | 31 | 18 | 3.145373 | 49 | P < 0.01 |
| Anaerotruncus | genus | BiomeSight | 2476 | 1835 | 2.82706 | 650 | P < 0.01 |
| Bacteroides | genus | uBiome | 283855 | 247858 | 2.683065 | 471 | P < 0.01 |
| Bifidobacterium | genus | uBiome | 30859 | 14357 | 2.650187 | 474 | P < 0.01 |
| Blautia | genus | BiomeSight | 109238 | 86161 | 3.171679 | 667 | P < 0.01 |
| Butyrivibrio | genus | Thryve | 2238 | 863 | 2.78185 | 414 | P < 0.01 |
| Chromatium | genus | BiomeSight | 54 | 23 | 2.899706 | 89 | P < 0.01 |
| Chryseobacterium | genus | Thryve | 112 | 28 | 5.423269 | 18 | P < 0.001 |
| Clostridium | genus | uBiome | 9065 | 7003 | 2.64738 | 469 | P < 0.01 |
| Clostridium | genus | BiomeSight | 24536 | 18253 | 3.27004 | 667 | P < 0.01 |
| Cronobacter | genus | uBiome | 7047 | 250 | 3.67629 | 51 | P < 0.001 |
| Desulfomonile | genus | BiomeSight | 94 | 34 | 3.874419 | 94 | P < 0.001 |
| Desulfovibrio | genus | uBiome | 8575 | 4552 | 2.939696 | 243 | P < 0.01 |
| Granulicatella | genus | uBiome | 839 | 137 | 2.901195 | 179 | P < 0.01 |
| Haemophilus | genus | uBiome | 12156 | 3087 | 3.235353 | 314 | P < 0.01 |
| Henriciella | genus | Thryve | 386 | 176 | 2.642494 | 155 | P < 0.01 |
| Hungateiclostridium | genus | BiomeSight | 4457 | 2097 | 2.785968 | 200 | P < 0.01 |
| Limosilactobacillus | genus | uBiome | 48109 | 2843 | 2.818423 | 44 | P < 0.01 |
| Macrococcus | genus | BiomeSight | 1558 | 388 | 2.673296 | 222 | P < 0.01 |
| Marvinbryantia | genus | uBiome | 4529 | 2564 | 2.773658 | 369 | P < 0.01 |
| Negativicoccus | genus | BiomeSight | 1370 | 264 | 3.038088 | 353 | P < 0.01 |
| Odoribacter | genus | uBiome | 8724 | 5405 | 3.26612 | 369 | P < 0.01 |
| Pelotomaculum | genus | BiomeSight | 259 | 96 | 2.808263 | 236 | P < 0.01 |
| Rothia | genus | Thryve | 2787 | 326 | 2.925505 | 153 | P < 0.01 |
| Rubritalea | genus | BiomeSight | 118 | 51 | 3.025228 | 232 | P < 0.01 |
| Senegalimassilia | genus | Thryve | 2823 | 617 | 3.184808 | 143 | P < 0.01 |
| Shuttleworthia | genus | Thryve | 105 | 75 | 2.606498 | 318 | P < 0.01 |
| Slackia | genus | uBiome | 3566 | 1488 | 4.3098 | 154 | P < 0.001 |
| Streptococcus | genus | uBiome | 12882 | 6211 | 3.438454 | 465 | P < 0.001 |
| Trabulsiella | genus | BiomeSight | 2381 | 72 | 2.944962 | 84 | P < 0.01 |
| Chryseobacterium group | norank | Thryve | 112 | 28 | 5.423269 | 18 | P < 0.001 |
| unclassified Parabacteroides | norank | uBiome | 2321 | 760 | 3.29071 | 91 | P < 0.01 |
| unclassified Streptococcus | norank | uBiome | 9530 | 3840 | 3.188989 | 429 | P < 0.01 |
| Bifidobacteriales | order | uBiome | 44064 | 18404 | 3.118635 | 363 | P < 0.01 |
| Chitinophagales | order | Thryve | 94 | 69 | 2.824838 | 330 | P < 0.01 |
| Desulfovibrionales | order | uBiome | 10291 | 6987 | 2.794631 | 401 | P < 0.01 |
| Eubacteriales | order | BiomeSight | 614126 | 553568 | 2.710308 | 667 | P < 0.01 |
| Flavobacteriales | order | Thryve | 377 | 93 | 4.138507 | 241 | P < 0.001 |
| Micrococcales | order | Thryve | 1469 | 345 | 2.755253 | 326 | P < 0.01 |
| Pasteurellales | order | uBiome | 11259 | 3343 | 2.952883 | 325 | P < 0.01 |
| Rhodocyclales | order | uBiome | 888 | 327 | 2.799376 | 44 | P < 0.01 |
| Rhodospirillales | order | uBiome | 27884 | 17148 | 3.002822 | 241 | P < 0.01 |
| Syntrophobacterales | order | BiomeSight | 75 | 45 | 2.874101 | 331 | P < 0.01 |
| Chloroflexi | phylum | Thryve | 1529 | 170 | 3.281628 | 262 | P < 0.01 |
| Cyanobacteria | phylum | uBiome | 6444 | 268 | 4.338711 | 12 | P < 0.001 |
| Fibrobacteres | phylum | Thryve | 180078 | 34 | 4.876895 | 83 | P < 0.001 |
| Firmicutes | phylum | BiomeSight | 646426 | 582627 | 2.769104 | 667 | P < 0.01 |
| Spirochaetes | phylum | Thryve | 22731 | 196 | 2.747053 | 68 | P < 0.01 |
| Adlercreutzia equolifaciens | species | BiomeSight | 549 | 211 | 3.480457 | 447 | P < 0.001 |
| Alistipes putredinis | species | uBiome | 14784 | 10219 | 3.567818 | 346 | P < 0.001 |
| Alistipes sp. NML05A004 | species | uBiome | 2861 | 1594 | 3.211406 | 270 | P < 0.01 |
| Anaerofustis stercorihominis | species | BiomeSight | 85 | 45 | 2.937001 | 130 | P < 0.01 |
| Anaerolinea thermolimosa | species | BiomeSight | 30 | 18 | 2.73704 | 47 | P < 0.01 |
| Anaerotruncus colihominis | species | BiomeSight | 2335 | 1749 | 2.696412 | 650 | P < 0.01 |
| Bacteroides finegoldii | species | BiomeSight | 4441 | 2045 | 2.613824 | 552 | P < 0.01 |
| Bacteroides heparinolyticus | species | BiomeSight | 59 | 33 | 2.652157 | 173 | P < 0.01 |
| Bacteroides nordii | species | uBiome | 5809 | 1153 | 3.833993 | 148 | P < 0.001 |
| Bacteroides reticulotermitis | species | Thryve | 724 | 301 | 4.303933 | 341 | P < 0.001 |
| Bacteroides sp. 35AE37 | species | uBiome | 24271 | 10025 | 3.724252 | 219 | P < 0.001 |
| Bacteroides uniformis | species | Thryve | 39266 | 26321 | 3.311011 | 405 | P < 0.01 |
| Bifidobacterium dentium | species | uBiome | 18963 | 288 | 3.177668 | 33 | P < 0.01 |
| Bifidobacterium longum | species | uBiome | 21851 | 6860 | 4.131341 | 271 | P < 0.001 |
| Bifidobacterium pseudocatenulatum | species | uBiome | 34144 | 8671 | 3.752826 | 94 | P < 0.001 |
| Blautia glucerasea | species | BiomeSight | 2184 | 606 | 2.887145 | 550 | P < 0.01 |
| Blautia obeum | species | BiomeSight | 11262 | 5088 | 4.539525 | 642 | P < 0.001 |
| Chromatium weissei | species | BiomeSight | 54 | 23 | 2.899706 | 89 | P < 0.01 |
| Corynebacterium spheniscorum | species | uBiome | 12700 | 3296 | 2.737118 | 99 | P < 0.01 |
| Desulfohalotomaculum peckii | species | Thryve | 35 | 16 | 3.191543 | 21 | P < 0.01 |
| Desulfomonile tiedjei | species | BiomeSight | 94 | 34 | 3.874419 | 94 | P < 0.001 |
| Granulicatella adiacens | species | uBiome | 626 | 100 | 2.637126 | 176 | P < 0.01 |
| Haemophilus parainfluenzae | species | uBiome | 10429 | 3084 | 2.943034 | 310 | P < 0.01 |
| Klebsiella oxytoca | species | BiomeSight | 2975 | 337 | 2.885423 | 113 | P < 0.01 |
| Negativicoccus succinicivorans | species | BiomeSight | 1381 | 248 | 2.970997 | 330 | P < 0.01 |
| Pelotomaculum isophthalicicum | species | BiomeSight | 259 | 95 | 2.82543 | 236 | P < 0.01 |
| Phocaeicola coprophilus | species | BiomeSight | 18667 | 2437 | 3.75374 | 151 | P < 0.001 |
| Phocaeicola plebeius | species | uBiome | 104385 | 39611 | 2.841081 | 49 | P < 0.01 |
| Porphyromonas bennonis | species | uBiome | 9563 | 2189 | 2.832604 | 170 | P < 0.01 |
| Ruminococcus bromii | species | BiomeSight | 15277 | 7865 | 2.709478 | 558 | P < 0.01 |
| Shuttleworthia satelles | species | Thryve | 102 | 71 | 2.89508 | 304 | P < 0.01 |
| Slackia piriformis | species | Thryve | 1676 | 634 | 2.804074 | 126 | P < 0.01 |
| Slackia piriformis | species | uBiome | 5996 | 1513 | 4.050816 | 41 | P < 0.001 |
Comments from Early Reviewers
“Interesting the connection between histamine and iron. I have some Mast Cell issues which have finally been diagnosed. I also have low ferritin [a blood protein that contains iron], although hemoglobin and even serum iron are within range…. BTW, I recall Hawrelak saying once that histamine behavior of bacteria is strain dependent, not species.”
3 thoughts on “Histamine Release – Literature Review And Speculation”
Comments are closed.