By same data, I mean the same FASTQ files, a detail file of the parts of your sample returned by a 16s machine. This is then processed through software to infer the bacteria. The result is two different reports. If you pass the same files to other providers, you will like get even more different reports. For why, see this post from 2019, The taxonomy nightmare before Christmas…
This post is going to look an actual example.

Krona View
At this level, they look similar – but there is often a 25% difference between the numbers of a species.
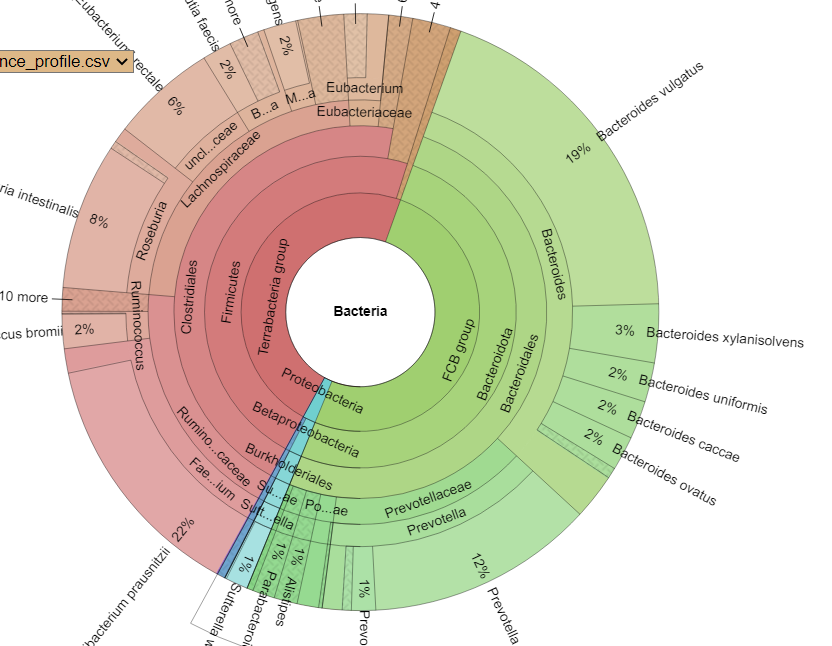
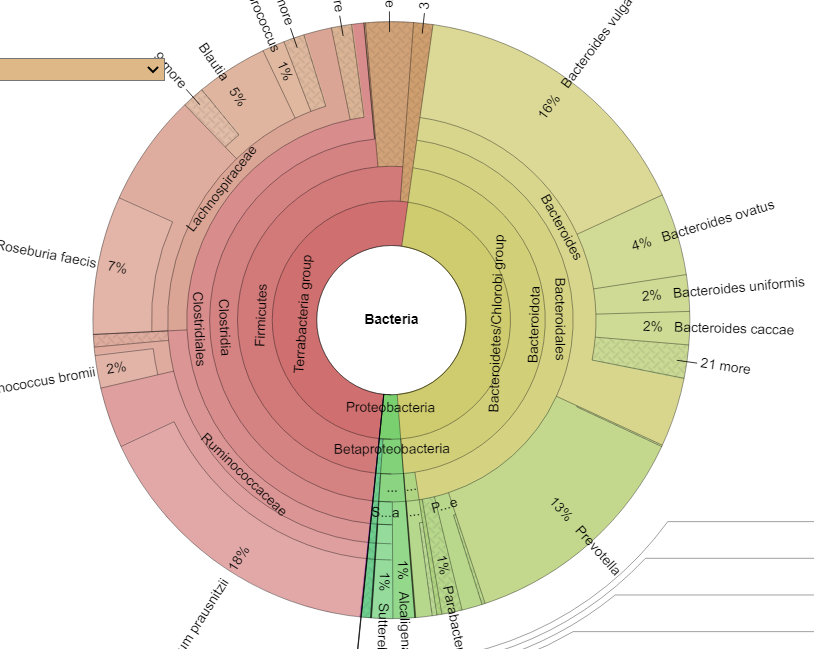
Comparing Samples
At the class level you can see some dramatic changes in counts and percentile. At present, I am using percentiles from aggregations of all labs sources.
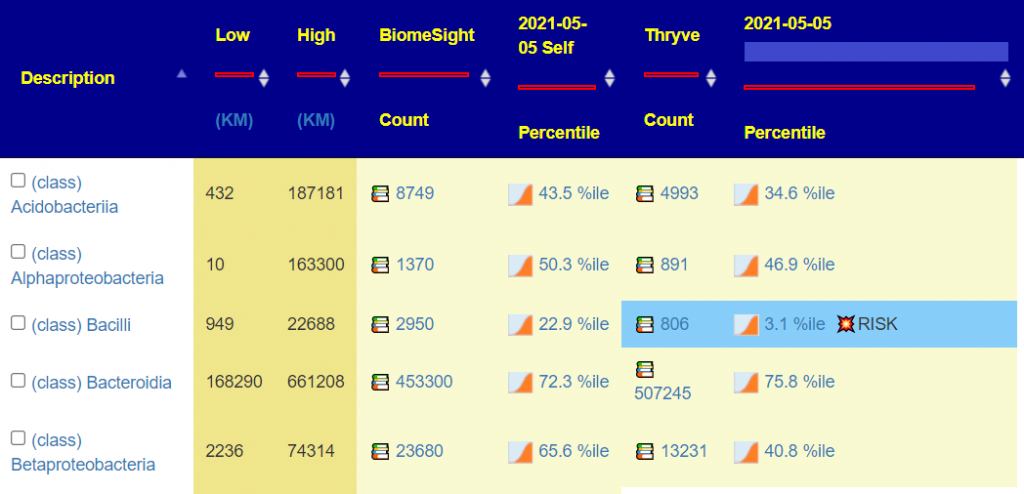
When I hit 1000 samples from a specific lab, I will doing lab specific percentiles. Current counts — thus we are using an aggregate for percentile for all labs
For items of concern, you can actually drill down manually on the bacteria. For example for Bacilli above.
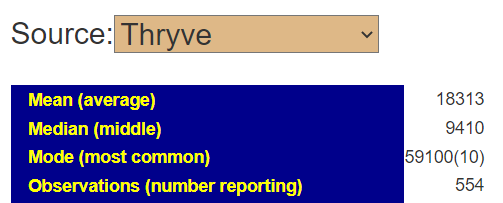
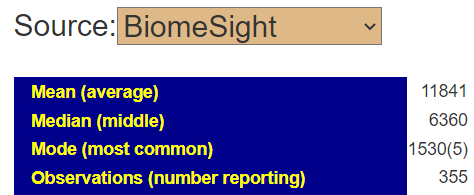
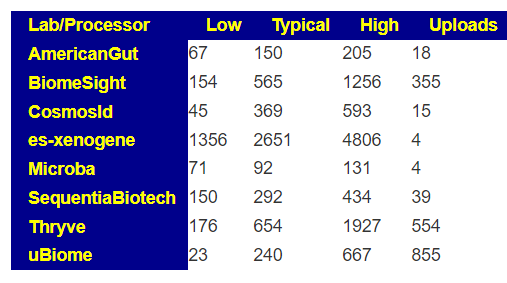
You can also get the percentile that is lab specific by going to https://microbiomeprescription.com/Library/Statistics?taxon=91061 with no sample and then changing to the lab as shown below.
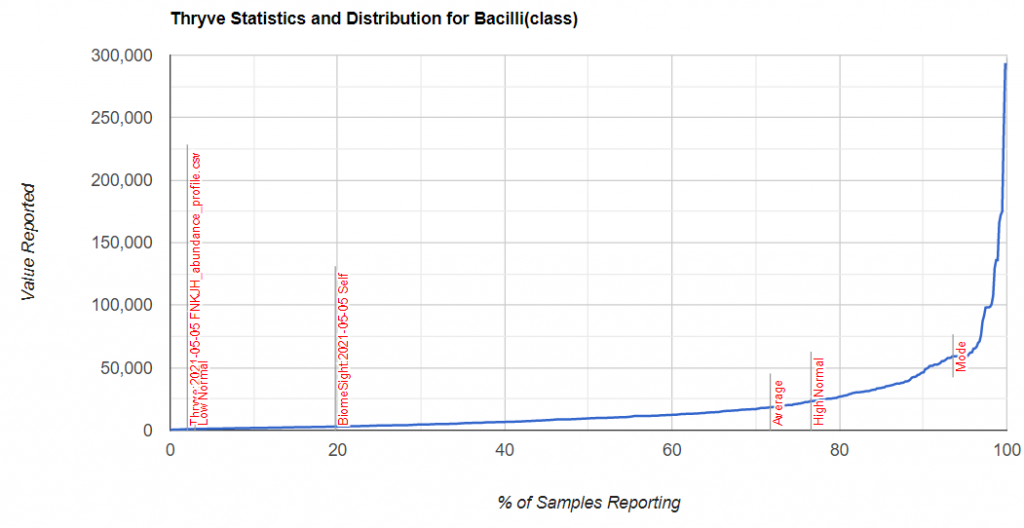
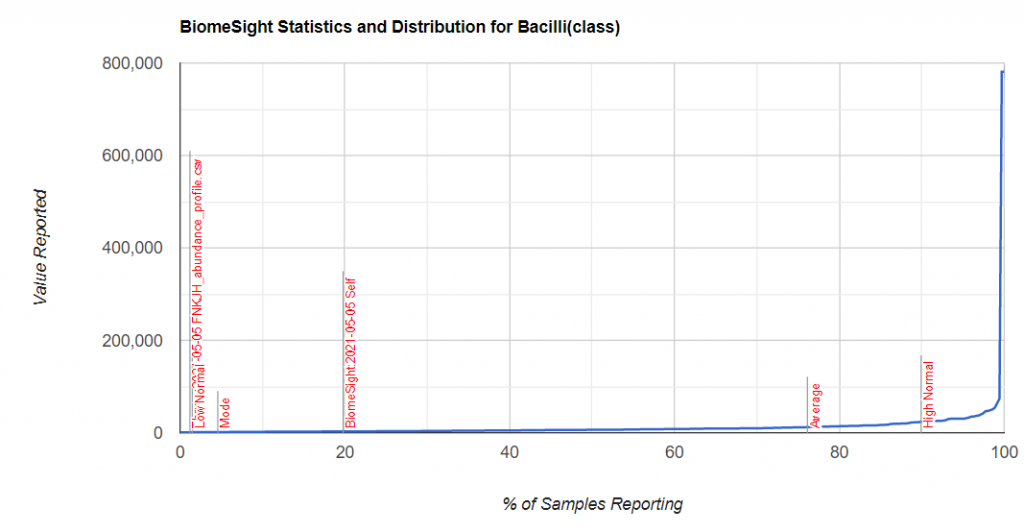
We find that we are at the 20%ile for biomesight specific samples and 2.4%ile for thryve specific samples. For explanations, you will need to ask the questions to the lab — microbiome prescription just presents the data.
Kegg Probiotic Suggestions
Different input present different outputs.
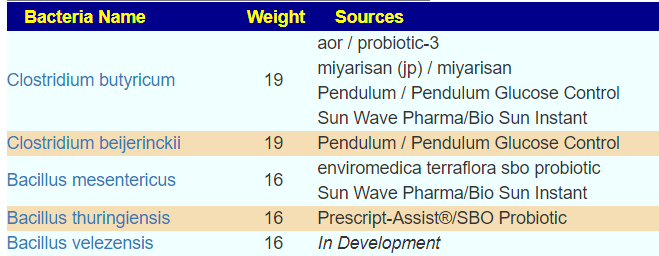
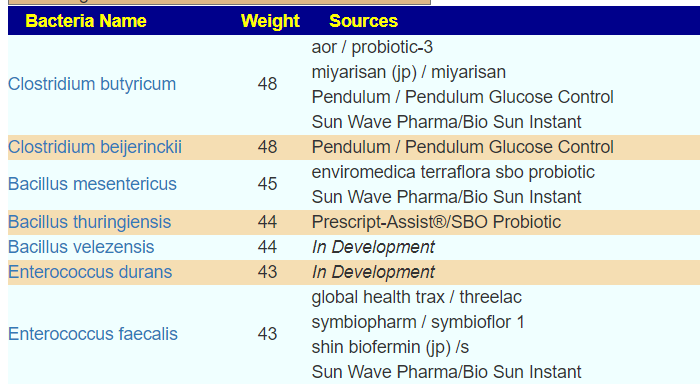
Dr. Jason Hawrelak Recommendations
One reports 9 items that are not ideal, and the other 8. There is disagreement on Blautia, Desulfovibrio, Lactobacillus, Roseburia and Bilophila wadsworthia
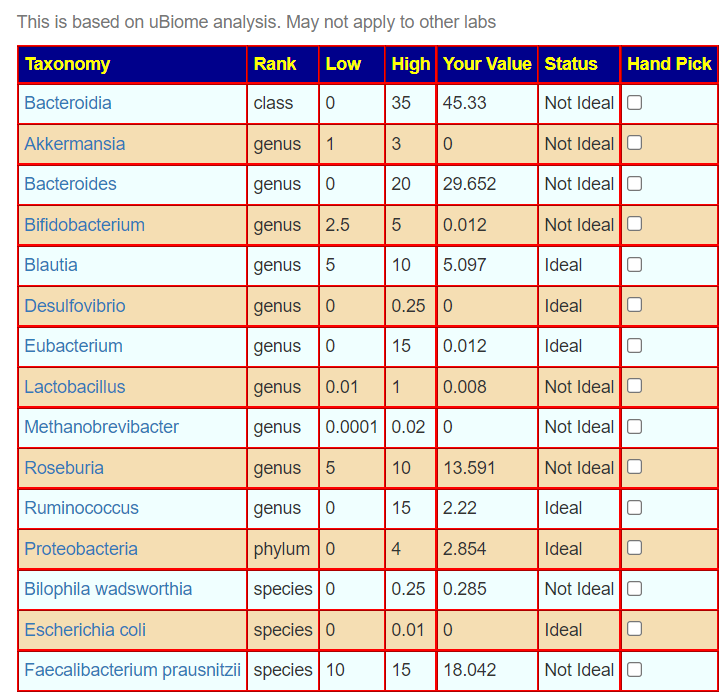
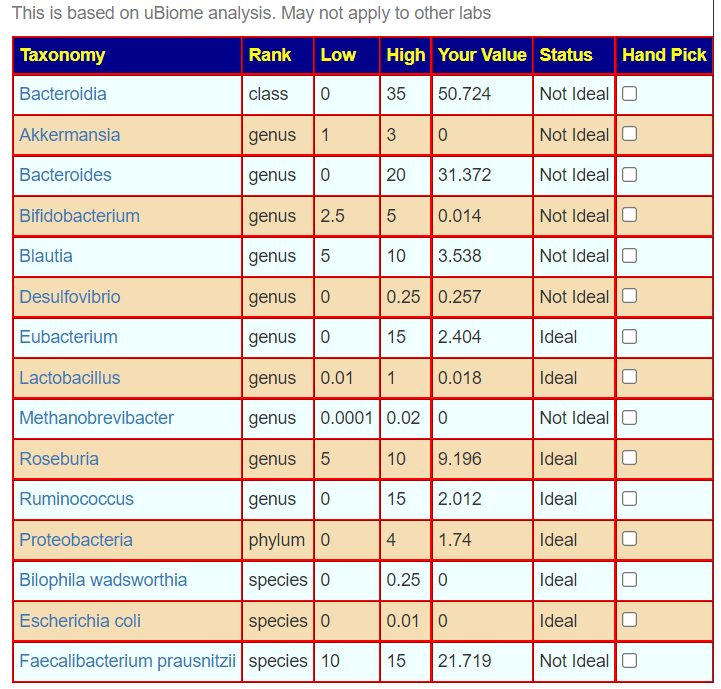
Bottom Line — FRUSTRATION!
The bottom line is that you want to always use the same lab software for comparing samples. Ideally, the same lab for the physical processing. Comparing the same sample that is processed by two different pieces of software results interpretation challenges.
To give a more human context — take a book and ask two people to retell it aloud, one is from the rural areas of Scotland (with thick Scottish accent) and the other from Mumbai India (with thick Marathi accent) with a third person (a native from Bermuda) trying to recall what they heard…. Different choice of words in the retelling with different intonations. That is the human reality — which also applies to labs.
Recent Comments