Microbiome Prescription has a deep database of facts on the microbiome. Most other sites lack any depth, in some cases almost none.
Traditional Approach
A microbiome testing firm will hire a dietician or health care provider to provide the information. This is known as “handing off to the experts”. These typical experts will use knowledge from courses they took, open a text book, and if you are lucky, do a few quick scans of the internet.

There are problems with this approach. Some simple examples: articles and studies retractions; new literature not seen. The purpose of this page is to show one approach that can be done to get better results if you use a moderately skilled software developer.
Microbiome Prescription Approach
Microbiome Prescription (MP) is expert system focused and thus focused on massive facts databases. The traditional approach typically produces “sound bites” that is often single or a few bacteria focused.
I use the US National Library of Medicine as my primary resource. This has a rich API that can be programmed against. For an example, consider autism.
Step 1 Get Studies IDs
First query ask for studies on autism, I restricted it to 10 simplicity
https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=autism&retmode=json&retmax=10

Note that it automatically does synonyms for you – SWEET!
“to”: “\”autism’s\”[All Fields] OR \”autisms\”[All Fields] OR \”autistic disorder\”[MeSH Terms] OR (\”autistic\”[All Fields] AND \”disorder\”[All Fields]) OR \”autistic disorder\”[All Fields] OR \”autism\”[All Fields]”
Step 2 Get Actual Studies
With the IDs above, we just drop them into a query like below. I just did 39569070,39568799:
https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=pubmed&id=39569070,39568799&retmode=xml
I did XML here because it presents better. The result has all of the data seen on PubMed, including the Abstract.
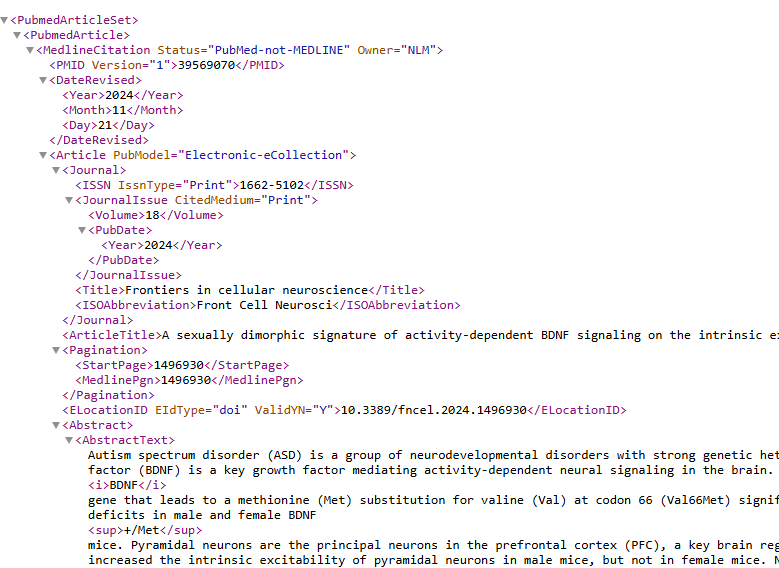
Step 3 Get Text To Use
If the abstract is sufficient we are done. If not, you can get the links for the full text from the above information.
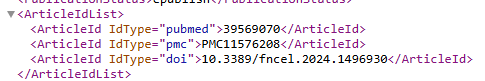
Step 4 Parse the text to identify key words
This means a little coding. Python on JSON works very well and is common with most younger developers. You need to select the terms to identify. I usually parse all documents and then show the results. Information should be stored in some form of efficient datastore. For example MongoDB’s popularity include:
- Expressive query language: Provides powerful indexing and aggregation capabilities
- Document data model: Allows for easy storage and combination of various data types.
- Dynamic schema: Enables quicker iteration and less time spent on data preparation.
Below you see the Conditions, Modifiers and Bacteria that I scanned for. After each there is a count of the number candidates PMIDs.
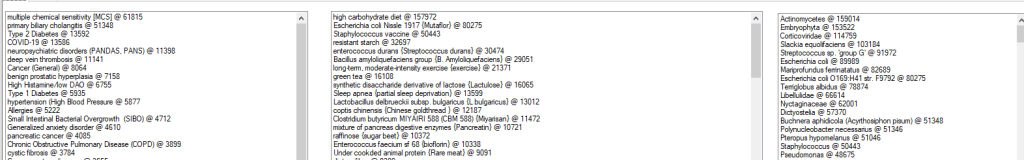
Storing all of this data on the cloud is strongly recommended because it can greatly speed processing.
Step 5 Examining a Study
This is pretty simple, I click on one of the above items and the page refreshes to show the studies (ordered by those with most items identified at the top). In this, we see just 300 studies
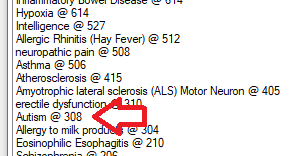
The result shows the text and the words and phrases match.
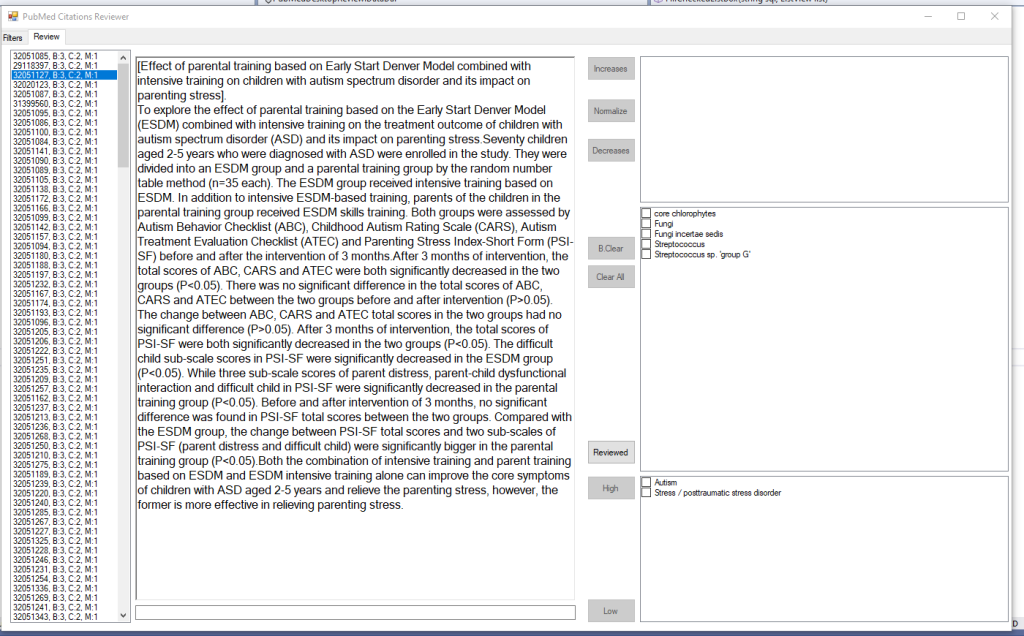
Then it is a fast reading of the citations and then checking the boxes and clicking a button
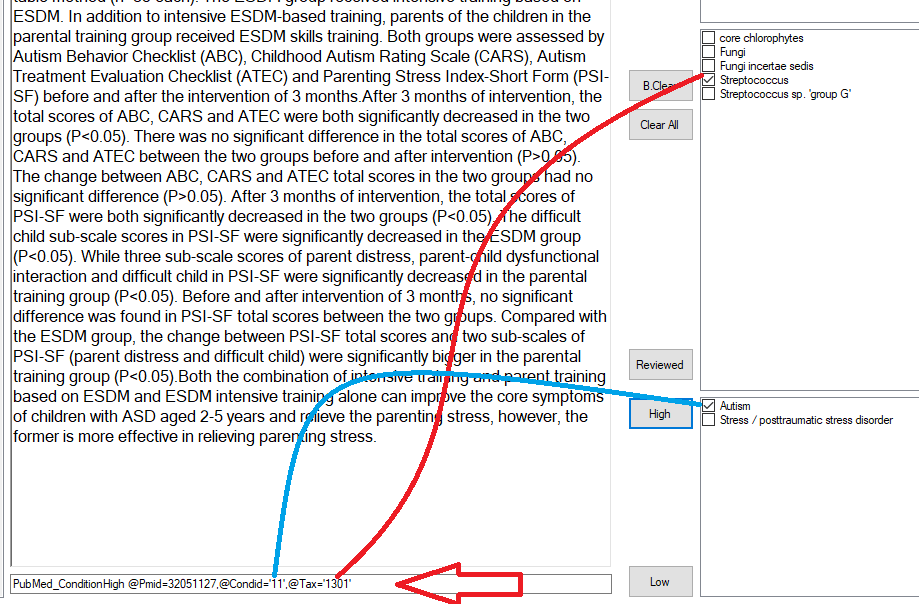
In this case, I showed the resulting command (to help triaging issues). When I am done with one study, I click [Reviewed] and it is removed from the pending review list. In a more enhanced version, I color code the bacteria names for faster visual parsing. Example below

Of course, there will be some false detections — which can be improved with tuning the parsing.
After a day of tuning(literally) and playing with the UI to speed the review, I produced the video below showing it’s use. Note that I added addition buttons to capture if a modifier helps or hurts a condition according to the study. This allows my cross-validated suggestions to expand to more conditions. Often you will see data that is valuable, there, but not captured. When practical, capture is a efficient manner.
Getting data from Pubmed for use in Expert Systems / AI
Bottom Line
The above process is heavily automated but with a final human review. A rate of 3-6 abstract review per minute is viable, If you are working with the full texts, this number will drop dramatically.
Recent Comments