Let us start with a more real world example: Dogs.
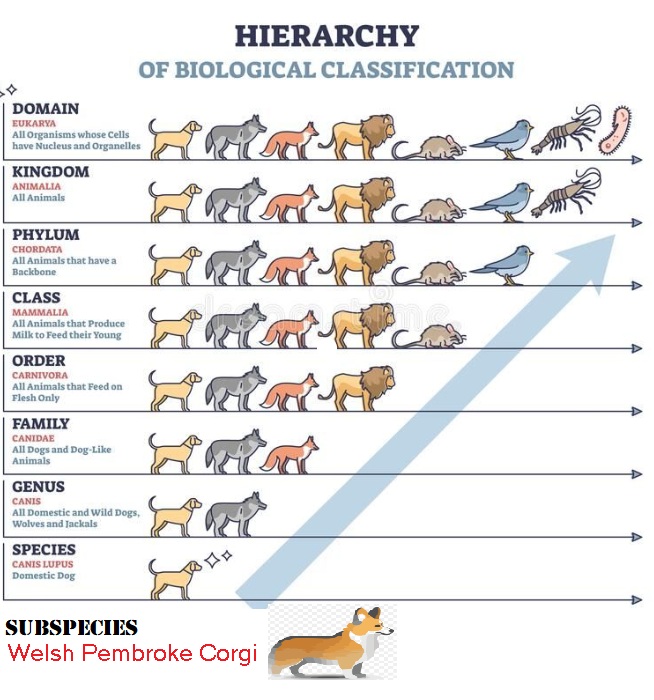
Take a vaccine against Rabies tested on dogs in a pound (Canis Lupis). It was successful. Inference means that there is a high probability that it would work for Welsh Pembroke Corgis — although there was none in the pound. This is a child inference.
There is a high probability that this vaccine would also work for the Genus Canis, which include wild dogs such as Jackals (Africa), Wolves, Coyote and Dingos (Australia). This is a parent inference.
There is a reasonable probability that this vaccine would also work for the Family Canidea which includes Foxes. This is a grandparent inference.
The key thing to remember is that each layer of the taxonomy hierarchy has significant DNA shared with those above and below. It is likely (not guaranteed) that the layer above or below will respond similarly.
A Common Inference Seen with Medical Consultants
A consultant may read an article like “Whole genome sequencing of Lacticaseibacillus casei KACC92338 strain with strong antioxidant activity…” and based on this study recommend Lactobacillus Casei probiotic for a patient. This is a parent inference. We do not know definitely if this general species would have any of the desired behavior. There is a reasonable probability. If you reject inference then you can only recommend this explicit strain, no substitutions allowed. If you are using herbs, Greek Oregano (Origanum vulgare L. ssp. hirtum) may be cited in the study (Origanum vulgare ssp. hirtum (Lamiaceae) Essential Oil Prevents Behavioral and Oxidative Stress Changes… so Oregano Oil cannot be assumed to do similar — that is an inference.
The Microbiome has stricter overlaps than mammals
In the last 20 years, different bacteria has been sequenced resulting in a more correct hierarchy based on DNA. For example, Lactocaseibacillus casei was originally Bacillus casei, then Lactobacillus casei. A short table of a few others is shown below.
| Current name | New name |
| Lactobacillus casei | Lacticaseibacillus casei |
| Lactobacillus paracasei | Lacticaseibacillus paracasei |
| Lactobacillus rhamnosus | Lacticaseibacillus rhamnosus |
| Lactobacillus plantarum | Lactiplantibacillus plantarum |
| Lactobacillus brevis | Levilactobacillus brevis |
| Lactobacillus salivarius | Ligilactobacillus salivarius |
| Lactobacillus fermentum | Limosilactobacillus fermentum |
| Lactobacillus reuteri | Limosilactobacillus reuteri |
We do not do sibling inference. Studies on Limosilactobacillus fermentum are not inferred to Limosilactobacillus reuteri, we do parent inference to Limosilactobacillus with no inference to Levilactobacillus, Lactiplantibacillus, Lactobacillus, nor Lacticaseibacillus (i.e. uncle inferences).
The recent reorganization of the bacteria hierarchy based on DNA makes inferences more probable.
Avoiding Inferences
It is technically possible to avoid inferences for some bacteria. For other bacteria, for example Propionibacterium freudenreichii subsp. shermanii, you may find just one study and that decreases only — when you want to increase it! Looking at Propionibacterium freudenreichii and inferences, you have over thirty studies. We do not know if these substances will work. There is a good probability that it may work
“Who you gonna Call? Call Sparse Data Busters!”
 | Using inference allows us to get suggestions with a reasonable chance of working. We give direct citations a high weight. We give inferences a diminished weight. Microbiome Prescription works off probability estimators when using inference. |
It’s your choice on Microbiome Prescription
Using inference is the user’s choice. You may agree or disagree on inference — if you disagree than please be consistent and only use the strains of probiotics cited in studies.
Recent Comments