In my prior post on ubiome Json files, or taxon number based files, I gave code and the pattern to use. Unfortunately, not every one provides such easy to use data. Look at the following download file format (AmericanGut)
#taxon relative_abundance k__Bacteria;p__Bacteroidetes;c__Bacteroidia;o__Bacteroidales;f__Bacteroidaceae;g__Bacteroides 0.287722 k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__;g__ 0.130347 k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Ruminococcaceae;g__ 0.116602 k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Ruminococcaceae;g__Faecalibacterium 0.045869 k__Bacteria;p__Bacteroidetes;c__Bacteroidia;o__Bacteroidales;f__Porphyromonadaceae;g__Parabacteroides 0.042162 k__Bacteria;p__Bacteroidetes;c__Bacteroidia;o__Bacteroidales;f__Rikenellaceae;g__ 0.039537 k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Lachnospiraceae;g__ 0.034286
We have the name hierarchy (which unfortunately can vary slightly by lab). The names identify the tax_rank (which can also vary by lab!).
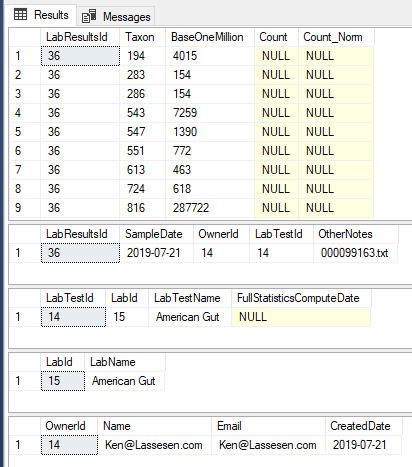
For this console application, I am going to end up with a data table consisting of:
- tax_rank (converted from f__, s__ etc)
- tax_name (extracted from the last name before the number)
- BaseOneMillion (done by multiplying the number by 1,000,000)
Source: https://github.com/Lassesen/Microbiome2/tree/master/taxonNameUpload
This data table is then uploaded and Sql goes thru a series of steps to attempt to match it. Things that are not matched are written to a file (like done in the prior post). In this case, the solution is much easier to patch the differences. Just add the name to the TaxonNames file with it’s appropriate taxon (no need to create a separate substitution table). The Sql attempts to match by taxon rank, name first, if that fails, it falls back to name alone.
For the test file that I used, there was a lot of mismatches

Spot checking for a few, I discovered that these are currently listed as having a parent of “unclassified Gammaproteobacteria” (118884)
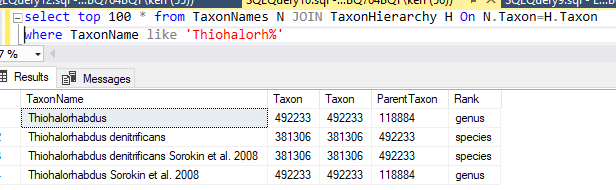
For others, there is nothing that seems to be a match and the original data line does not clarify anything.
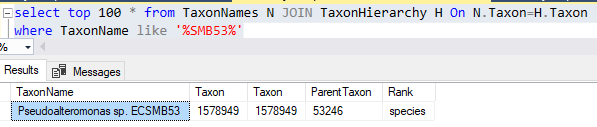
k__Bacteria;p__Firmicutes;c__Clostridia;o__Clostridiales;f__Clostridiaceae;g__SMB53 0.000927
Bottom Line
For a 103 line input we got 87 matches and the rest unclear. The sample code gives you the information on the challenges. You need to come up with your own resolutions (discard or patch)
P.S. Remember to update your SQL Server Schema to get the new sprocs and data types.
1 thought on “Upload Taxon Name based Files”
Comments are closed.