For many months, using R2 Associations to select probiotics used a generic database from PrecisionBiome. Over the year end holidays, I computed the R2 Associations based on data from specific labs. This is far more accurate and have just been added for the following labs:
- Biomesight (best because biggest dataset)
- Ombre
- uBiome
- Thorne (smallest dataset and not as much data).
At the bottom of the suggestion page you will see a new section like below:
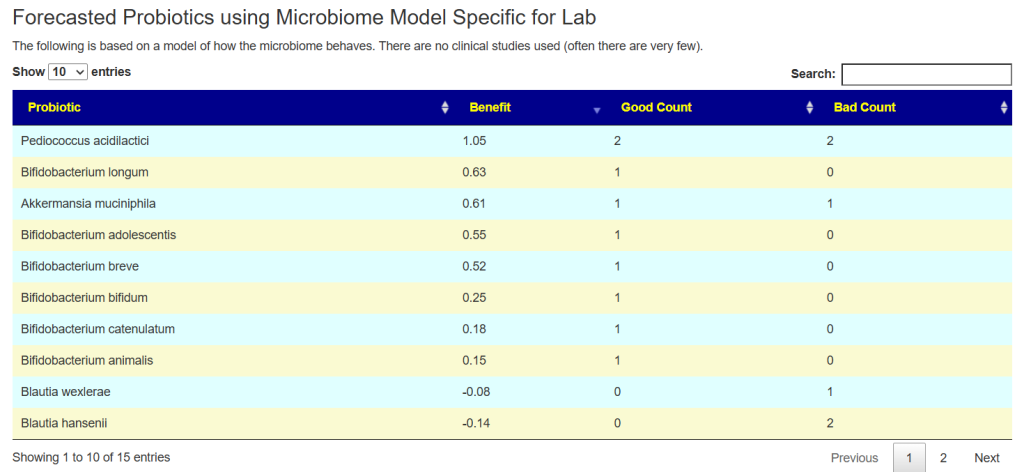
The range of numbers can vary greatly.
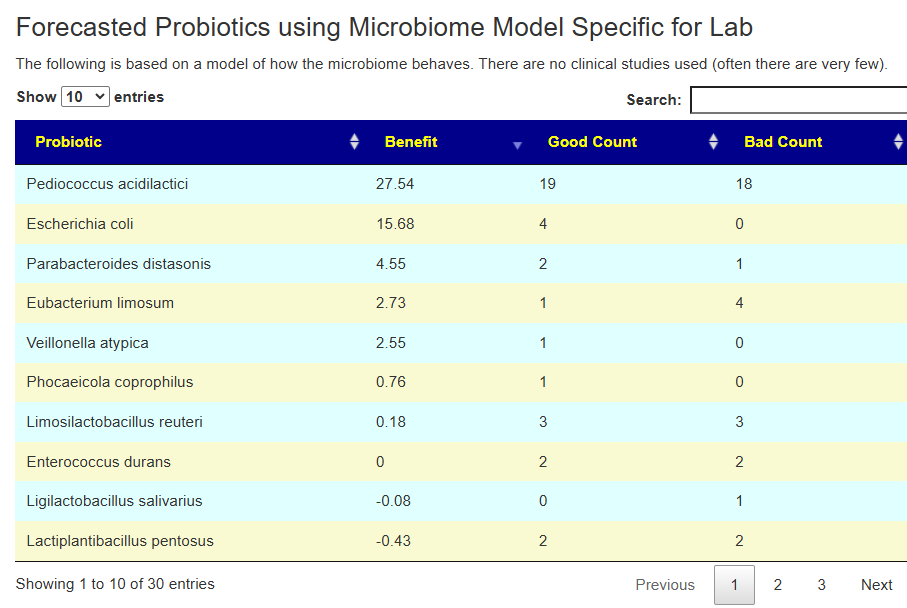
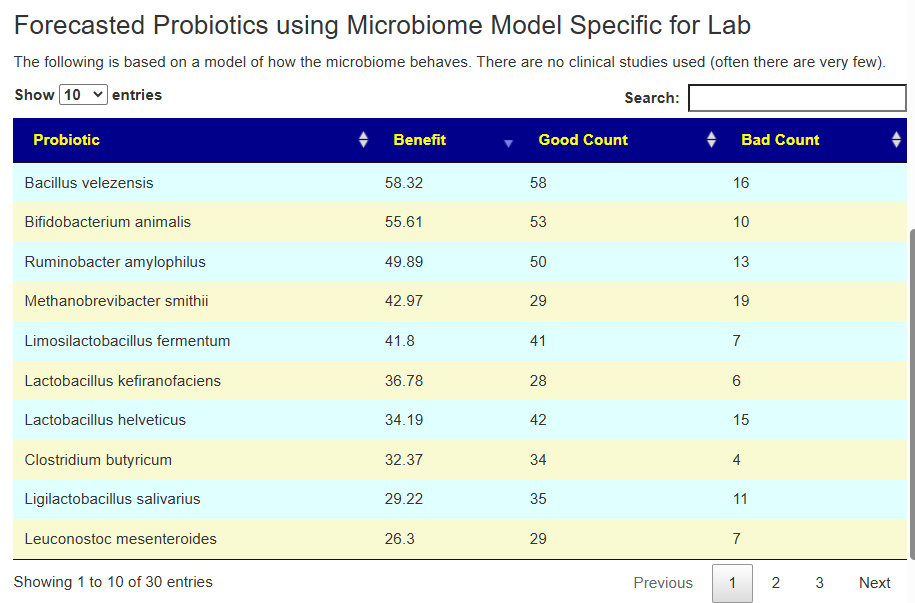
Which is best? We have multiple ways of computing probiotics. The ideal would be to do only ones that each way advocates. When there are Good and Bad counts, having a positive good and zero for bad is ideal.
The reality is that this rarely happens. I tend to favor Good count much bigger than Bad with a high positive benefit. Our knowledge is sparse and often studies results fail to duplicate. I tend to favor this method because it is an Fuji apple to Fuji apple comparison instead of the Crab Apple to Watermelon comparison that published studies tend to be.
Recent Comments