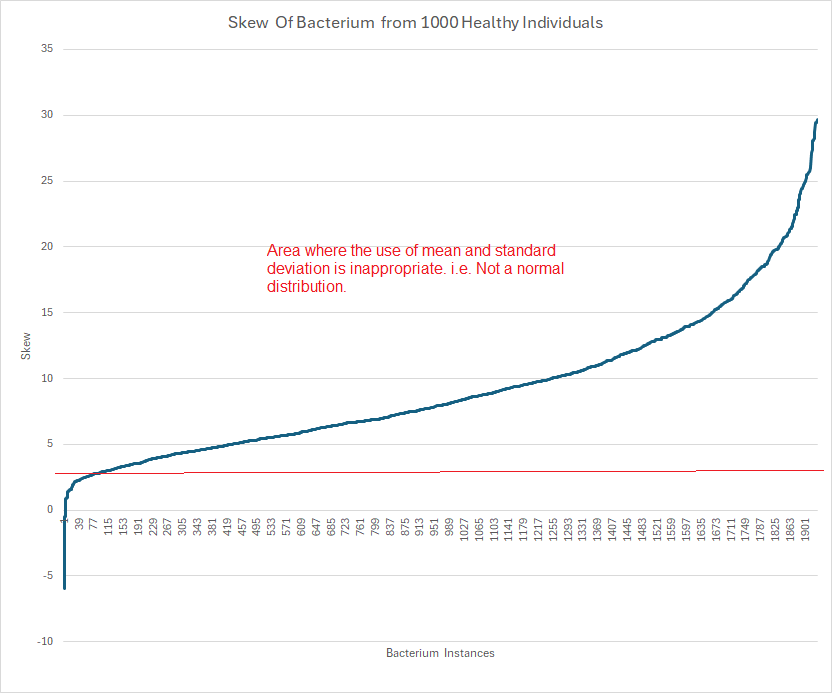
In an earlier post (Significant Bacteria and Their Thresholds – Part 1), I raised that issue and a EU colleague, Valentina Goretzki, suggested that I take data from 1 thousand shotgun samples from healthy individuals to illustrate the problem.
Microbiome data distributions frequently display extreme skewness—often greater than 20. In such cases, computing mean and standard deviation is simply incorrect. My friend “Perplexity” writes Mean and standard deviation become inappropriate measures for computing significance if the distribution’s skewness is substantial—specifically, when the absolute skewness exceeds ±2.
The result was about two thousand bacterium that occurs at least 60 times in these samples could be plotted as shown below.
It is clear that non-parametric methods are needed to compute “healthy ranges”. For those with just basic statistics, this may become a significant challenge.
Recent Comments