Anyone who regularly reads peer-reviewed medical studies on the microbiome will notice findings reported as bacteria being “too high” or “too low,” with phrases like “trending” when statistical significance isn’t reached. Frankly, my reaction to 95% of these papers is an eye-roll, as the statistical methods used are often inappropriate for the data at hand. With multiple degrees in statistics, professional memberships, and experience, I’m acutely aware of both best practices and common pitfalls.
Microbiome data distributions frequently display extreme skewness—often greater than 20. In such cases, computing mean and standard deviation is simply incorrect. My friend “Perplexity” writes Mean and standard deviation become inappropriate measures for computing significance if the distribution’s skewness is substantial—specifically, when the absolute skewness exceeds ±2.
Despite this, using these metrics remains standard in high school statistics and unfortunately persists in many life science studies. This “comfort zone” approach does nothing but cloud true findings in microbiome science.
My alternative methodology uses a much larger, highly annotated dataset—over 4,290 unique samples generously donated to Microbiome Prescription., most transferred from Biomesight.com. Importantly, these samples are uniformly processed and richly annotated with symptoms rather than diagnoses, yielding superior analytical clarity.
My Natural Questions to ask
Natural for a statistician that is.
- For people with a symptom or diagnosis
- What are the significant bacteria associating (and likely causing) the symptom
- What is the threshold levels for these bacteria
- I use levels and not level because I have observed the same symptom may occur with a bacteria outside of a specific range. That is, too high or too low. I have also encountered this reported in a few studies, often hidden under a term like “altered microbiome”.
- There is a dangerous assumption in the literature that significant bacteria must be either too high or too low. I unfortunately know Kierkegaard’s “Either/Or” well.
- There are no universal threshold for all symptoms, each has its own
- For people without a symptom with a statistical model but with dysbiosis
- How do you determine which bacteria are significant?
- What is the threshold levels for these bacteria
Over the last decade, these are important questions because they lead directly to treatment suggestions.
They are also significant in evaluating progress. At present I have a forecasting algorithm that has a high prediction rate for symptoms from a microbiome. The forecasting algorithm also is useful for evaluating progress, an example for a recent sample the donor asked me to review.
Prediction
The checks indicates that the donor agrees that they have this symptom.
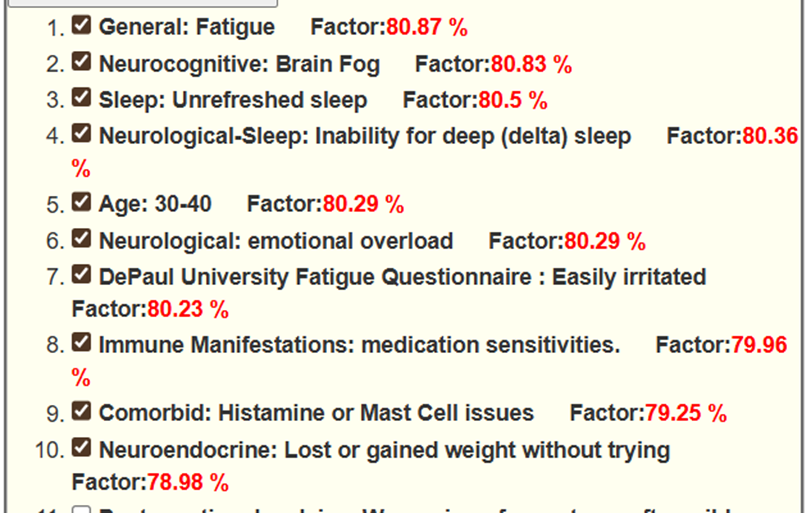
Monitoring
The person above followed the suggestions and the subsequent test results are shown below.
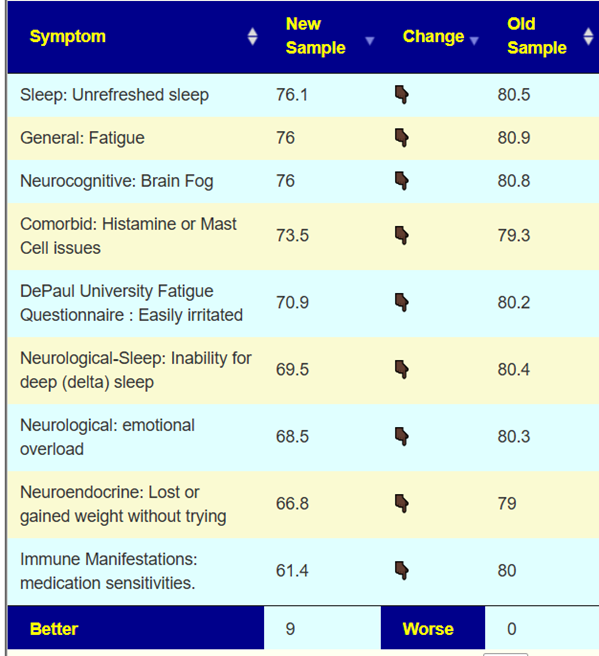
What are the most common bacteria associated with symptoms?
This is a generic question that is useful for health practitioners to know. For example, Kristina Mitts, of Mind Mood Microbiome and who I frequently correspond with, or Dr. Jason Hawrelak.
Using more appropriate statistical methods on our sample of 4,292 distinct different samples; we found significant bacteria identified over 327 symptoms resulting in the following statistical significances.
| Significance: P < | Count |
| 0.05 | 13,855 |
| 0.01 | 12,411 |
| 0.001 | 7,614 |
| 0.0001 | 5,532 |
So what are the top one for each of these significance?
Overall Significance
| Taxon | name | rank | Instances |
| 820 | Bacteroides uniformis | species | 165 |
| 35833 | Bilophila wadsworthia | species | 142 |
| 35832 | Bilophila | genus | 139 |
| 818 | Bacteroides thetaiotaomicron | species | 137 |
| 1426 | Parageobacillus thermoglucosidasius | species | 133 |
| 118884 | Gammaproteobacteria incertae sedis | no rank | 125 |
| 871324 | Bacteroides stercorirosoris | species | 124 |
| 120580 | Symbiobacterium toebii | species | 122 |
| 53244 | Desulfonatronovibrio | genus | 122 |
| 543349 | Symbiobacteriaceae | family | 122 |
| 2733 | Symbiobacterium | genus | 122 |
| 1498 | Hathewaya histolytica | species | 122 |
| 454155 | Paraprevotella xylaniphila | species | 120 |
P < 0.05
| Taxon | name | rank | Instances |
| 2950010 | Salidesulfovibrio | genus | 47 |
| 221711 | Salidesulfovibrio brasiliensis | species | 46 |
| 658623 | Chelonobacter | genus | 45 |
| 69224 | Erwinia psidii | species | 44 |
| 213462 | Syntrophobacterales | order | 44 |
| 3024408 | Syntrophobacteria | class | 44 |
| 31977 | Veillonellaceae | family | 44 |
| 1843489 | Veillonellales | order | 43 |
| 550 | Enterobacter cloacae | species | 43 |
| 35832 | Bilophila | genus | 42 |
| 841 | Roseburia | genus | 41 |
| 53244 | Desulfonatronovibrio | genus | 41 |
| 871324 | Bacteroides stercorirosoris | species | 41 |
| 1260 | Finegoldia magna | species | 41 |
| 1498 | Hathewaya histolytica | species | 40 |
P < 0.01
| Taxon | name | rank | Instances |
| 35833 | Bilophila wadsworthia | species | 51 |
| 78448 | Bifidobacterium pullorum | species | 50 |
| 820 | Bacteroides uniformis | species | 47 |
| 841 | Roseburia | genus | 46 |
| 818 | Bacteroides thetaiotaomicron | species | 44 |
| 118884 | Gammaproteobacteria incertae sedis | no rank | 41 |
| 1769729 | Hathewaya | genus | 41 |
| 1426 | Parageobacillus thermoglucosidasius | species | 41 |
| 112902 | Propionispora | genus | 40 |
| 36853 | Desulfitobacterium | genus | 40 |
| 386414 | Hoylesella timonensis | species | 40 |
| 119065 | unclassified Burkholderiales | family | 40 |
| 1853231 | Odoribacteraceae | family | 40 |
| 400091 | Hymenobacter xinjiangensis | species | 39 |
| 209080 | Propionispora hippei | species | 39 |
| 871324 | Bacteroides stercorirosoris | species | 39 |
| 69224 | Erwinia psidii | species | 39 |
| 35832 | Bilophila | genus | 39 |
P < 0.001
| Taxon | name | rank | Instances |
| 820 | Bacteroides uniformis | species | 50 |
| 35833 | Bilophila wadsworthia | species | 37 |
| 35832 | Bilophila | genus | 32 |
| 118884 | Gammaproteobacteria incertae sedis | no rank | 32 |
| 658623 | Chelonobacter | genus | 31 |
| 246787 | Bacteroides cellulosilyticus | species | 31 |
| 120580 | Symbiobacterium toebii | species | 31 |
| 543349 | Symbiobacteriaceae | family | 31 |
| 253238 | Ethanoligenens | genus | 31 |
| 2733 | Symbiobacterium | genus | 31 |
| 292833 | Candidatus Rhabdochlamydia | genus | 30 |
| 324707 | Candidatus Rhabdochlamydia crassificans | species | 30 |
| 1426 | Parageobacillus thermoglucosidasius | species | 30 |
| 689704 | Candidatus Rhabdochlamydiaceae | family | 30 |
| 70190 | Chroococcus | genus | 29 |
| 402401 | Chroococcus minutus | species | 29 |
| 1890464 | Chroococcaceae | family | 29 |
| 283169 | Odoribacter denticanis | species | 28 |
P < 0.0001
| Taxon | name | rank | Instances |
| 820 | Bacteroides uniformis | species | 40 |
| 246787 | Bacteroides cellulosilyticus | species | 34 |
| 818 | Bacteroides thetaiotaomicron | species | 30 |
| 1963360 | Parachlamydiales | order | 30 |
| 454155 | Paraprevotella xylaniphila | species | 30 |
| 1426 | Parageobacillus thermoglucosidasius | species | 30 |
| 2733 | Symbiobacterium | genus | 29 |
| 543349 | Symbiobacteriaceae | family | 29 |
| 120580 | Symbiobacterium toebii | species | 29 |
| 35832 | Bilophila | genus | 26 |
| 191412 | Chlorobiaceae | family | 25 |
| 256319 | Chlorobaculum | genus | 25 |
| 244127 | Anaerotruncus | genus | 25 |
| 189723 | Prevotella micans | species | 25 |
| 53244 | Desulfonatronovibrio | genus | 25 |
| 324707 | Candidatus Rhabdochlamydia crassificans | species | 24 |
| 191410 | Chlorobiia | class | 24 |
| 35833 | Bilophila wadsworthia | species | 24 |
Summary
This is a high level overview of Significant Bacteria. The patterns above are specific for tests done by Biomesight; a lack of standardization results in using these identifications for other tests is unsafe (legal sense). Background here. IMHO, it is a moral responsibility for labs to produce similar tables.
The key findings are:
- “Common suspects” such as bifidobacterium and lactobacillus are missing!
- Large sample sizes with the same processing is critical. The processing must be the same as used in a clinical setting.
- Appropriate statistical methods must be used
Stay tune for the next part as we drill deeper into appropriate handing of data with some specific issues like Long COVID.
The input data that I used is publicly available at: https://citizenscience.microbiomeprescription.com/
Post Script
Probiotics and the above gets interesting. Take Bacteroides uniformis which is at the top of many of these tables. If we go to my bacteria association site,
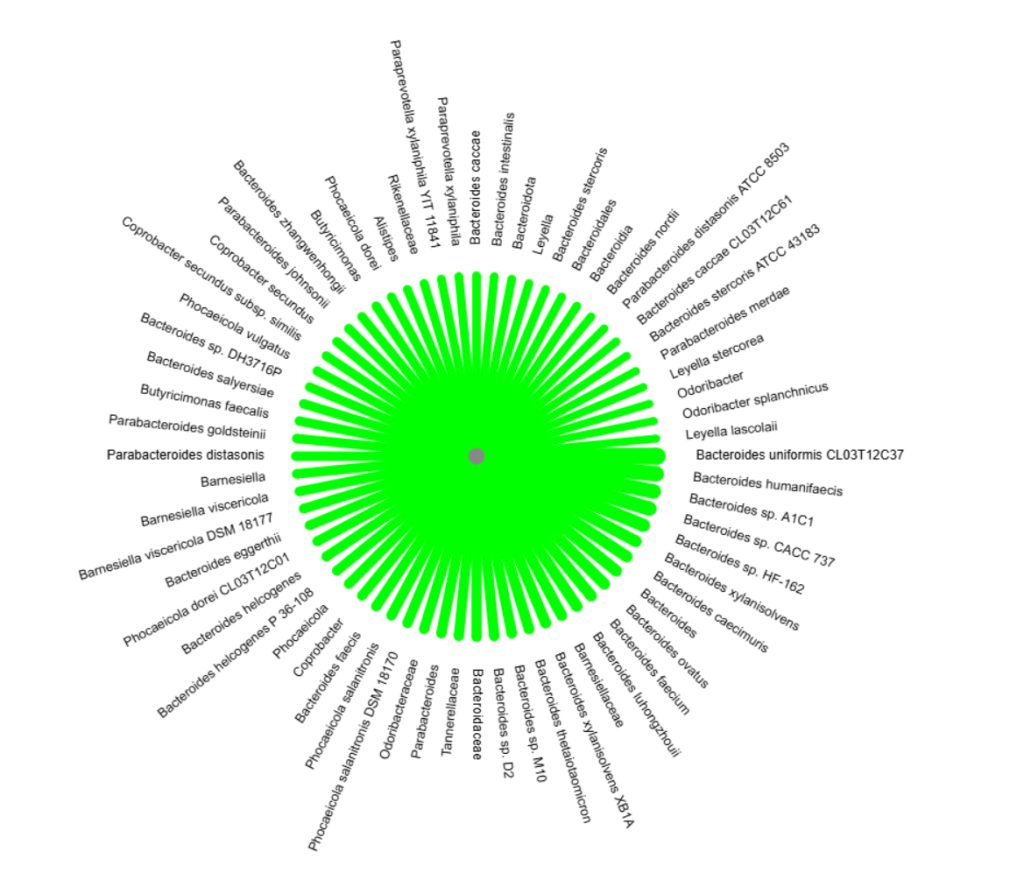
We can determine the probiotics (available or pending) that will increase this bacteria (none decreases)
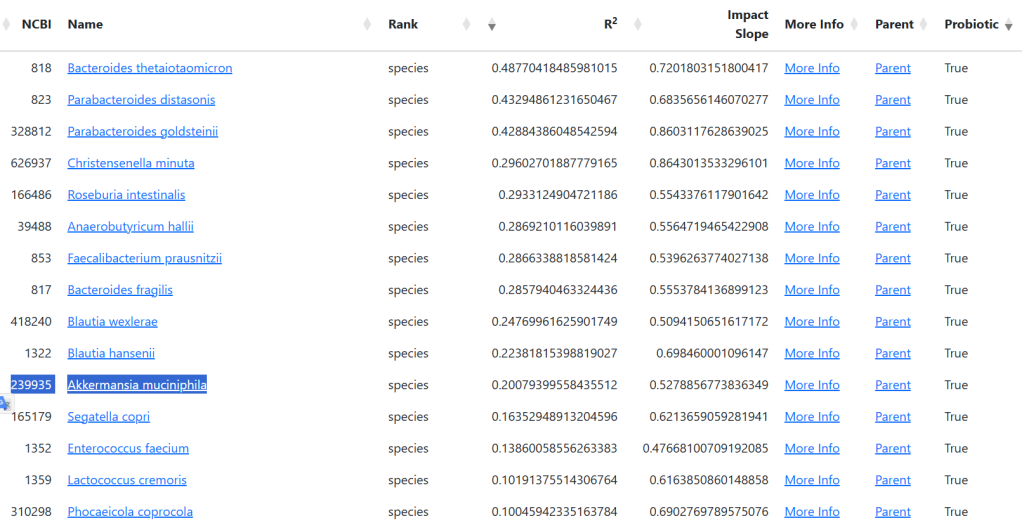
Again, the “cure all” lactobacillus and bifidobacterium genus is absent (apart from Ligilactobacillus ruminis which is not currently available).
Recent Comments