Related posts:
- Probiotics Fundamentals: Part 1 Specific Strains
- Probiotics Fundamentals: Part 2 DOA on Shelf
- Probiotics Fundamentals: Part 3 Are they working?
In the first post of this series, Probiotics Fundamentals: Part 1 Specific Strains I cited strains that are available retail that has been researched. The logical starting point is to search for your needs, read the studies and then rank the probiotics in prefer order for doing a personal trial. You want to do one probiotic at a time with rotation and described in the prior post (see prior post).
To searching for strain specific studies of probiotics available retail. Click here.
No Study found or issue not listed
The next step is to look at the conditions that I have abstracted/extracted studies for, listed at “U.S. Nat. Lib. Medical Conditions Studies with Microbiome Shifts“. We are shifting from strain to species level. This gives several paths, let us examine Autism. There are
Based on Publish Studies of Species
Clicking on [Can You Help Improve Suggestions] will take you to a page. At the bottom you will see “Treatment Substances” which lists things that have helped in studies. Scan it for probiotic names, for example:
A synbiotic formulation of Lactobacillus reuteri and inulin alleviates ASD-like behaviors in a mouse model: the mediating role of the gut-brain axis. Food & function (Food Funct ) Vol: 15 Issue: 1 Pages: 387-400 Pub: 2024 Jan 2 ePub: 2024 Jan 2 Authors Wang C,Chen W,Jiang Y,Xiao X,Zou Q,Liang J,Zhao Y,Wang Q,Yuan T,Guo R,Liu X,Liu Z
Which suggests L. Reuteri with inulin may help. The source is linked. Make sure you read them.
Based on Deficiencies of Probiotic Bacteria
Clicking on 🦠 Taxons will take you to a page showing all of the bacteria shifts reported for the condition.
Look for Lactobacillus, Bifidobacterium,etc with ⬇️
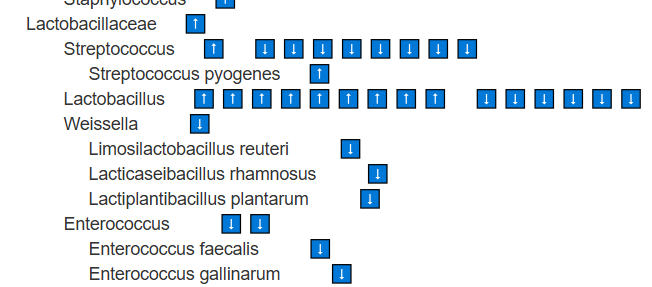
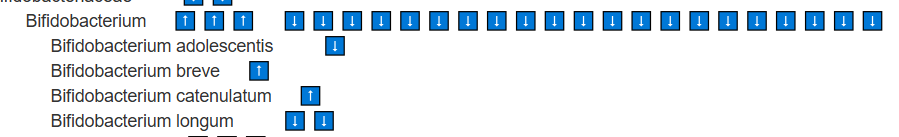
These species are found at lower levels, suggesting their metabolites are also reduced. Supplementing with them as single-strain probiotics is logical. Stay at the species level (e.g., Bifidobacterium longum) rather than higher classifications such as the genus Bifidobacterium. In general, avoid probiotic mixtures, as they may include strains that are counter-indicated (e.g., Bifidobacterium catenulatum, Bifidobacterium breve) or strains for which we lack sufficient information.
Based on Modelled correction of Bacteria
Clicking on 🥣 Candidates, will send the huge bacteria list above through a fuzzy logic expert system to compute suggestions with weights given for each one.
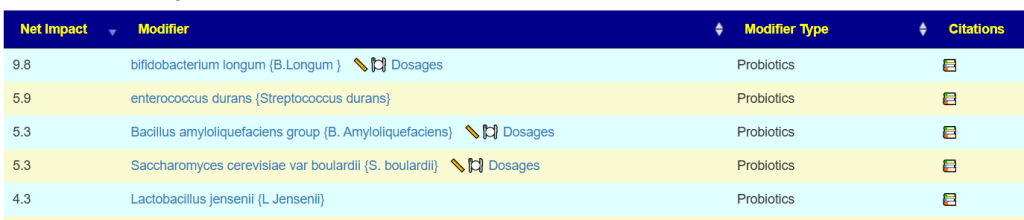
Note that this also lists ones to avoid.

Disagreements!
We can see that levels of Lactobacillus plantarum are low, but the model is telling us to avoid Lactobacillus plantarum. So what’s going on here?
The issue comes from the fact that the model/studies is based on multiple subgroups of people with Autism (or other conditions). The data might be accurate within each subgroup, but when you merge them together, you can end up with contradictions. So it’s not really a problem with the approach—it’s a problem with the data mix.
The best rule of thumb is to start with the things that show up as agreements across the data. For example: Bifidobacterium longum and Limosilactobacillus reuteri. Once you’ve tried probiotics that have clear agreements, then you can carefully experiment with the ones where there’s disagreement and see how your body responds.
The next level up in Probiotic Suggestions
It is pretty simple, get a microbiome test. My preferred tests are:
- Biomesight for 16s (economic, low resolution)
- Thorne for shotgun (more expensive but much higher detail)
You want to ideally get a test that reports on all of the common probiotic bacteria. Many common tests do not report many of these. For example: Diagnostic Solution GI-Map reports only
- Bifidobacterium [genus] and no species
- Lactobacillus [genus] and no species
On the other hand both of the above tests report species.
When you select a test, you should check Microbiome Prescription to see what the detection rate is. For example for Bifidobacterium longum, we see how often this is detected in samples.
- For the shotgun tests (Xenogene and Thorne) we see 96% and 100% of the time, if it show low, you can have confidence in taking some
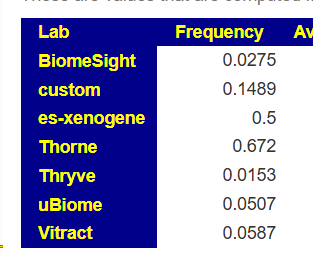
- For SequentiaBiotech we see it is seen 25% of the time. If you have none reported we are left being uncertain if you actually have none or is the none because of the test’s methodology
- See this post for more on these issues: The taxonomy nightmare before Christmas…
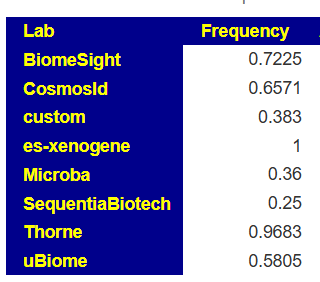
Another example is L.Reuteri where the shotgun tests find in in over 50% of samples, while some 16s finds in only 2% of samples.
Bottom Line
We’re piecing things together from lots of scattered knowledge, and there’s no single standard method—either for testing microbiomes in labs or for the studies themselves. Nothing here is clear-cut; everything’s kind of fuzzy, sometimes super fuzzy. In this post, the focus was on picking probiotics for a condition using literature (an “a priori” approach). Basically, it means trusting the data at face value, even though we know it isn’t rock-solid.
Some additional readings:
- Navigating microbiome variability: implications for research, diagnostics, and direct-to-consumer testing [2025]
- The standardisation of the approach to metagenomic human gut analysis: from sample collection to microbiome profiling [2022]
- Standardization of laboratory practices for the study of the human gut microbiome [2022]
I also foreshadowed the next post: Using a detailed microbiome test to select probiotics based on the whole microbiome.





Recent Comments