This is a post in this series !Technical Notes on Microbiome Analysis
The Question
A reader forwarded this study to me:
Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences [2018]
While this paper is dealing with fungi the tables can be eye opening for some people. A suitable quote from the paper “When the accuracy of genus predictions was averaged over a representative range of identities with the reference database (100%, 99%, 97%, 95% and 90%), all tested methods had ≤50% accuracy on the currently-popular V4 region of 16S rRNA.“
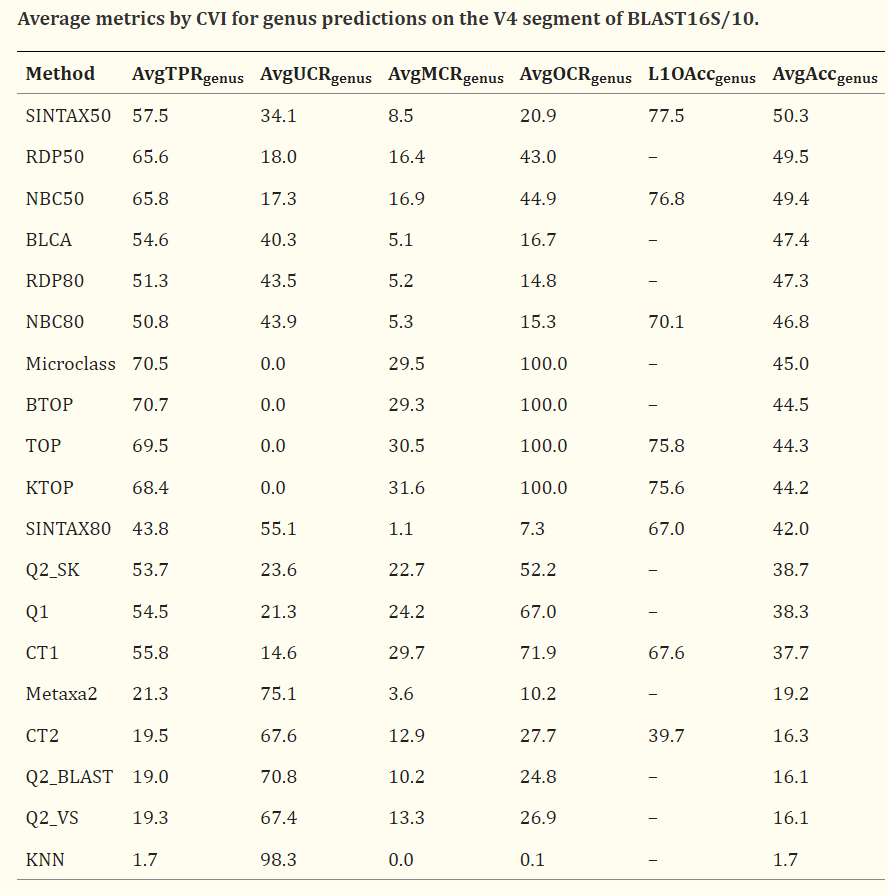
My expertise is in statistics, operational research and artificial intelligence, with good expertise in reading medical studies; so I asked a colleague who has a Ph.D. in Molecular Genetics. His casual comments were:
There are several studies with ASVs out there. Especially the recent ones. Clustering pipeline is what matters here. But I agree that full length gives better taxonomic assignment.
Problem is full length is twice as expensive. So my point is when using V4, you will achieve incredibly better taxonomic assignments with ASV vs OTU. However, full length or V3-V5 gives a better resolution.
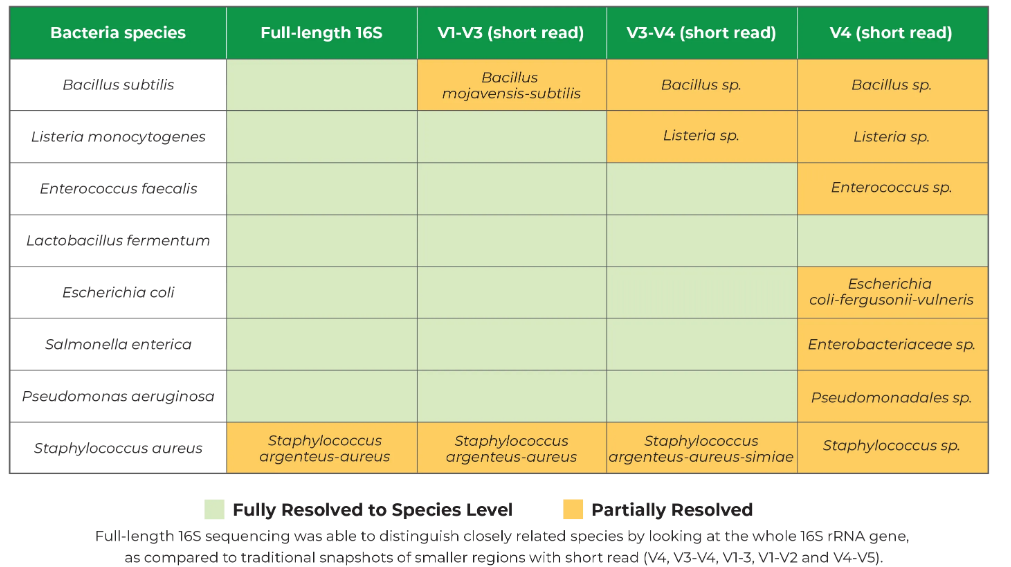
He also shared this graphic from Zymo Research. The V4 often cost around $50 and the full length can be 3-4x more.
What is ASV?
- ASV stands for amplicon sequence variants.
- OTU stands for operational taxonomic units
ChatGPT gives a good common man explanation:
Both methods aim to characterize and quantify the diversity of microorganisms in a given sample, but they differ in their underlying algorithms and conceptual frameworks.
- Amplicon Sequence Variants (ASVs):
- ASVs are derived from high-throughput sequencing data by clustering sequences that differ by as little as a single nucleotide. This means that ASVs are defined at a very fine level of sequence resolution.
- The goal of ASVs is to represent individual unique sequences within a dataset, thereby capturing the most detailed information about the microbial community present in a sample.
- ASVs are typically generated using algorithms like DADA2 (Divisive Amplicon Denoising Algorithm 2), which infer exact sequence variants and correct sequencing errors.
- ASVs are considered more accurate in capturing true biological diversity but may be more sensitive to sequencing errors.
- Operational Taxonomic Units (OTUs):
- OTUs are clusters of similar sequences that are defined based on a chosen sequence similarity threshold (commonly 97% similarity for bacterial 16S rRNA gene sequences).
- The 97% similarity threshold is often used to group sequences into OTUs to approximate the species level, although this can vary depending on the marker gene and research goals.
- OTUs are generated using methods such as UCLUST, UPARSE, or others that involve sequence clustering. The resulting OTUs represent a consensus sequence for each cluster.
- OTUs are considered more tolerant to sequencing errors, but they may group together closely related species or strains into the same cluster.
In summary, the main difference lies in the level of sequence resolution. ASVs aim for the highest possible resolution by identifying unique sequences, while OTUs represent clusters of similar sequences based on a chosen threshold. The choice between ASVs and OTUs depends on the specific research goals, the desired level of taxonomic resolution, and considerations related to sequencing error handling and computational resources.
To translate into human terms: ASV identifies criminals by fingerprints or DNA, while OTU identifies by the image from a security camera.
A Dilemma for Direct-To-Retail Tests
My colleague words makes the points clearly: Problem is full length is twice as expensive. Consumers are not knowledgeable about the differences but are very cost aware. The cheapest and least reliable way is often the norm. A direct to retail test costing less than $400 is likely to use the more inaccurate processes.
A Dilemma for Data from Studies
We encounter the same issue often for studies, budget! Searching the US National Library of Medicine for ASV, I get 2,955 results
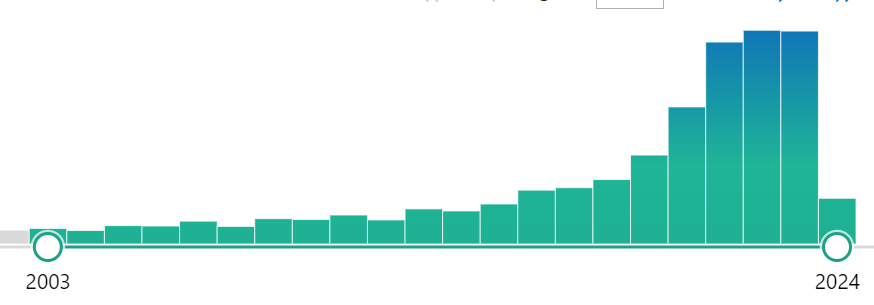
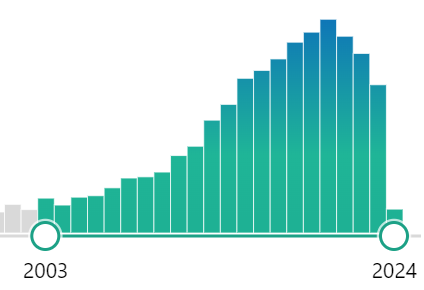
Searching the US National Library of Medicine for OTU, I get 9,180 results. We also see that ASV is replacing OTU starting around 2021.
This means that many studies published before 2021 may have correctly identified the bacteria impacted as little as 50% of the time. So, does Barley increases or decreases Bifidobacterium?
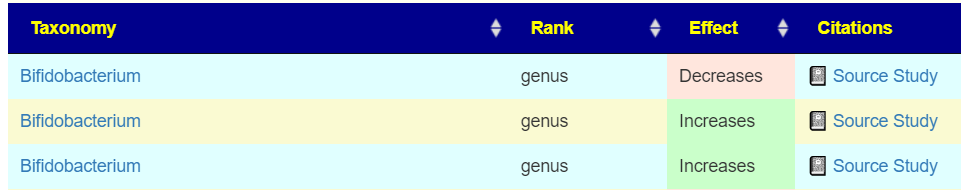
In addition to possible confounders with selection of control and subjects in the study, we must now consider the possibility of misidentification of the bacteria. For myself and microbiome prescription’s expert system, this is not a major issue because we are using a fuzzy logic expert system. Suggestions are based on most probable given the data available.
Many medical practitioners (MDs and naturopaths) are not trained in this area and resort to a naïve deterministic approach.
Additional Suggested Literature
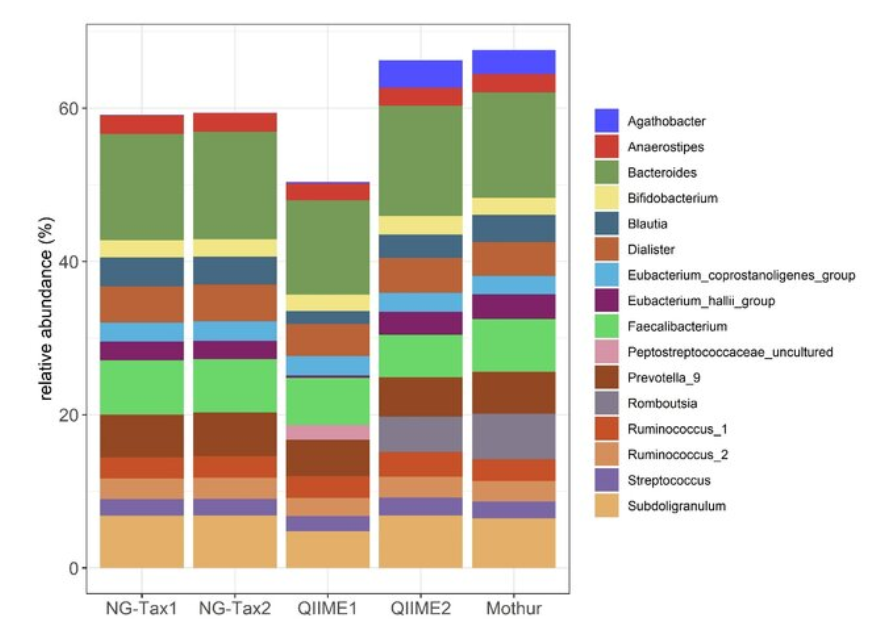
A comparison of bioinformatics pipelines for compositional analysis of the human gut microbiome [2023]
The differences of the same sample, Bacterial genera profile. Top 10 most abundant bacterial genera per pipeline resulted in a total of 16 unique genera.
- Additional recent work has shown that individual pipelines themselves may be biased toward certain phyla [15,21]
- The Illumina sequencing output reported an average quality of Q30 ≥ 81.9%.
- Based on mock communities, ASV-based approaches had a higher sensitivity in detecting the bacterial strains present, sometimes at the expense of specificity [17–20]
- OTUs detected much higher amounts of Verrucomicrobiae in the seston and sediment samples than were detected by the ASV approach. These differences are surprising given that both OTU and ASV approaches classified sequences to the same database.
Bottom Line
In dealing with microbiomes in a clinical setting, we have multiple fuzziness:
- The actual bacteria being reported (and the amount) is not reliable (in the common sense of that word), it is probable.
- When trying to modify the microbiome, the impact on the reported bacteria is not reliable (in the common sense of that word), it is probable.
This means using a single study has significant risk. With a diverse collections of studies and facts, then a fuzzy logic expert system results in significantly reduced risk and a higher probability of successful manipulation. It also illustrates why the Large Language Model (i.e. ChatGPT style) is very inappropriate. and likely machine learning also.
As of this writing, Microbiome Prescription has 10,390 Citations from US National Library of Medicine resulting in 2,415,340 facts in it’s expert system.
Recent Comments