A reader email me informing me that Microba no longer supplies 4 files, just one — Species. So no Phylum, Family or Genus information. I have modified the upload to handle just having one file available for whatever usefulness is possible.
A Long Time Pain
Microba does not provide data with the official NCBI Taxon numbers (which is what drives Microbiome Prescription AI). This means having to do name match over 3,635,527 different names on the NCBI database. Unfortunately, we still do not find matches, a few examples:
- Pauljensenia MIC9711
- QAMI01 MIC9451
- Ruminiclostridium_C sp000435295
- UBA1191 MIC6579
- UBA1191 MIC9444
Getting more information usually result in this from Google:

I have written them in the past hoping they would be more willing to cooperate. No luck.
What is Available
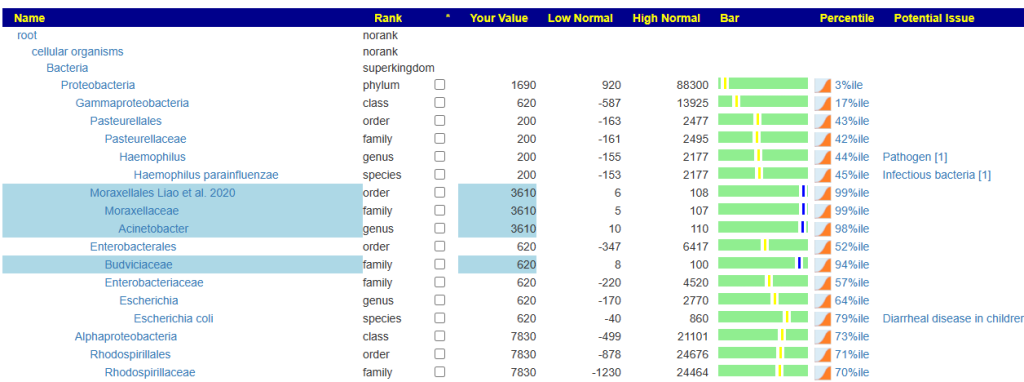
When you look at the microbiome tree, you will see values only for species. Everything else is 0. It is impossible to accurately estimate genus etc from species alone.
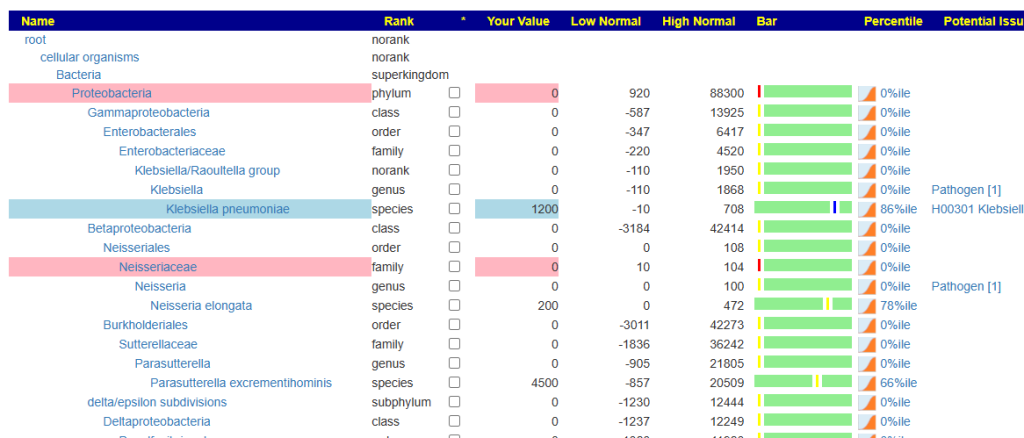
Prior, we would see something like this:
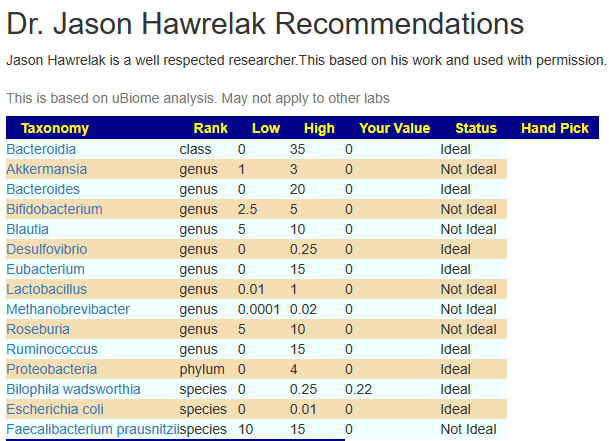
And you will get nothing from Dr. Jason Hawrelak Recommendations. Only one species is in it.
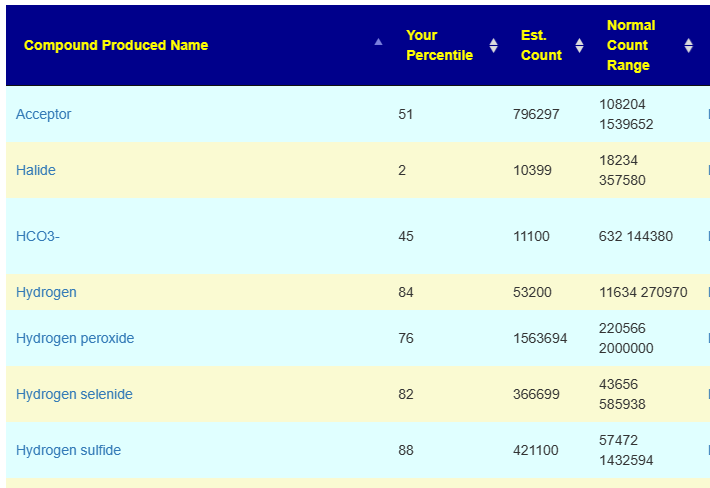
On the positive side, you will still get reasonably accurate KEGG based data (since that is based on species).
“Just Give Me Suggestions” will still work to some extent. Other suggestions can be tricky. The AI will work to the extent of the data available (which is greatly reduced compared to Thorne, Ombre or Biomesight).
Bottom Line
For those people in Australia, I would suggest moving over to BiomeSight, you will likely get six times more data at a lower cost.
Recent Comments