The data on Microbiome Prescription is based on studies and reports done using 16s methodology. At present, 16s studies greatly dominate reports on NIH National Library of Medicine. Similarly, ,major resources like KEGG: Kyoto Encyclopedia of Genes and Genomes and National Center for Biotechnology Information use NCBI Taxon numbers to clearly identified the bacteria.
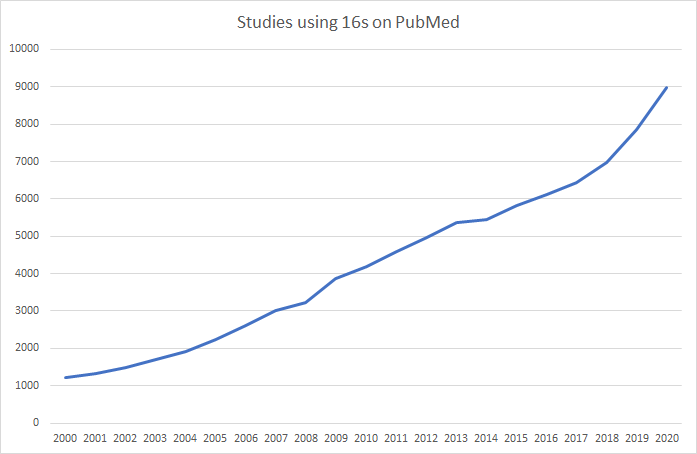
The key for creating an upload is identifying the taxon and the percentage of bacteria for a taxon.
Example
A reader wanted to upload his results and the lab would not provide a suitable CSV file. What they had was a page like shown below
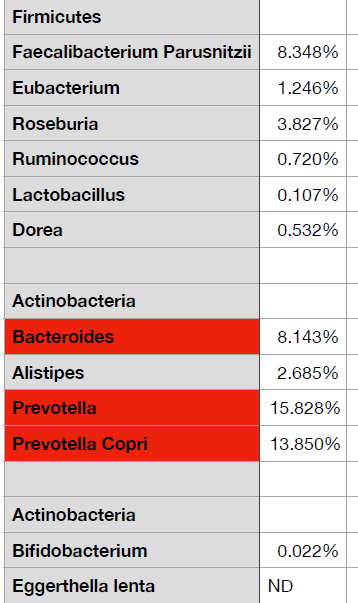
The starting point is simple, hand copy the data to Excel or equivalent (in many cases you can just copy the page or report and paste into Excel
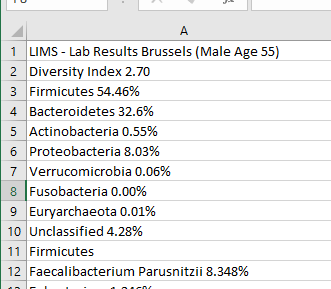
The next steps
- Delete any lines that do not have a measurement
- Copy the percentage (or compute it) into a new column.
For those familiar with excel I used these 3 formulas
- C: =FIND(” “,A3)
- D: =LEFT(A3,C3)
- E: =SUBSTITUTE(MID(A3,C3,10),”%”,””) * 1
The result is partial success with a few errors like below

For those caused by compound names, I need to count the characters to the last space and update Column C (I cheated by counting from the end and using =LEN(A25)-6. We ended up with
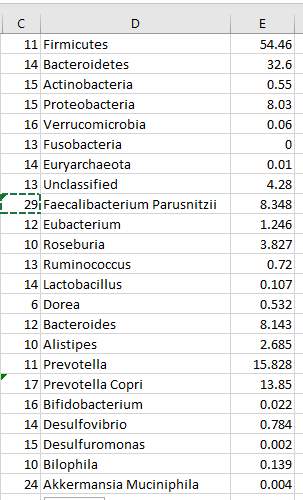
Also “Unclassified” should be deleted.
The Long Lookup
The next stage is time consuming… looking up the taxon numbers for each bacteria. There are two sources:
- Lookup on NCBI, i.e. https://www.ncbi.nlm.nih.gov/search/all/?term=lactobacillus
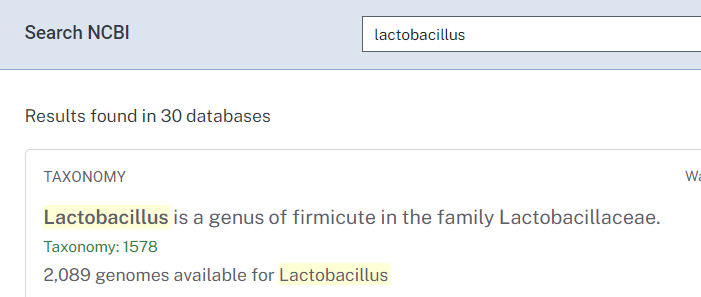
- Alternatively use https://microbiomeprescription.com/Library/Lookup
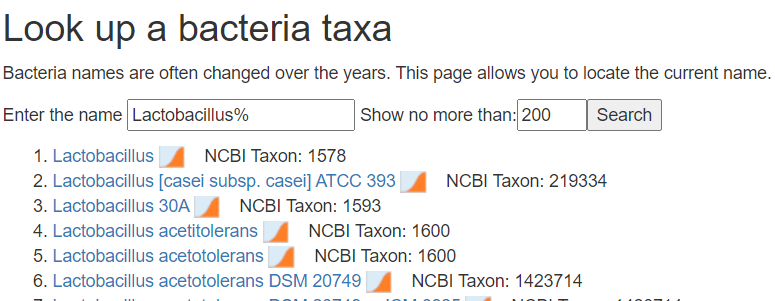
Add a new column before the percentage and put the taxon numbers there.
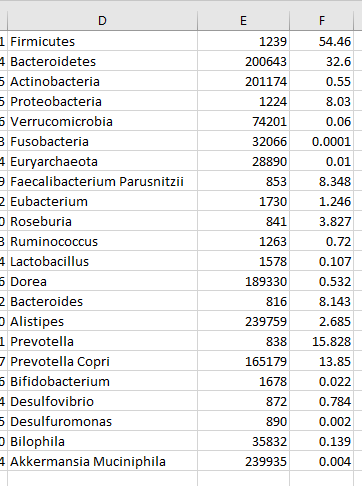
Next copy rows E and F to a new worksheet (remember to paste as values) and save as CSV file
The file should look like below.
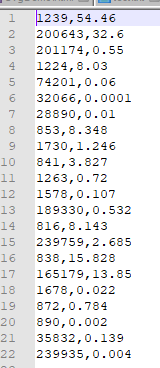
Now Insert your email address as the first line. resulting in:
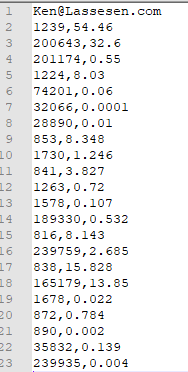
Next go to https://microbiomeprescription.com/Partners/transfer and paste the text there and click Test
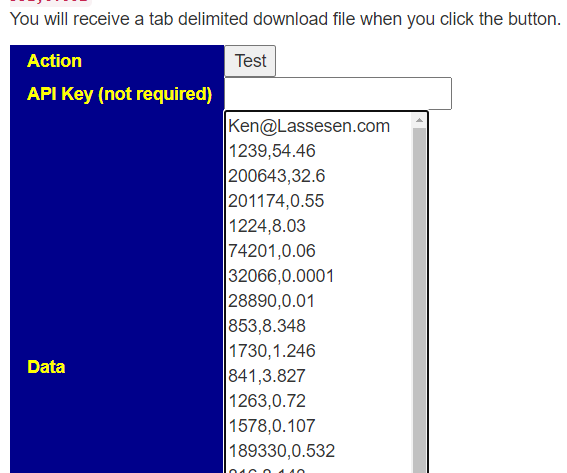
You will be prompted for a download:
Which will contain the details about the bacteria:
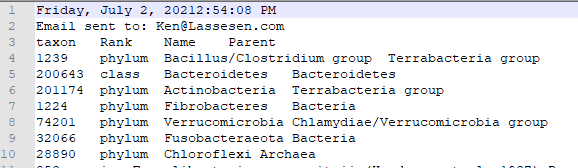
You will also receive an email

Getting it saved
After verifying that everything is correct…
It is simple — enter “custom” for the API Key — That’s it. The information will show up on your next login.
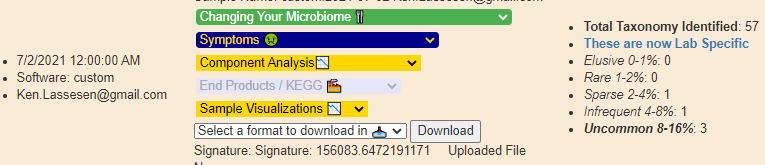
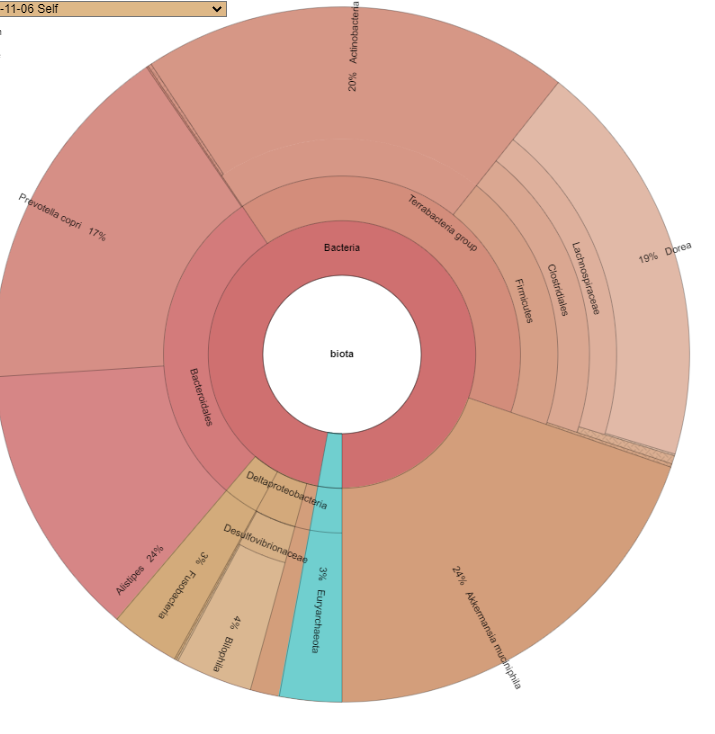
- Data submitted as custom is excluded from percentile and other computation — it is assumed to be incomplete
- The number of taxon will usually increase, the system automatically add layers of the hierarchy when missing, our 22 lines became 57 automatically
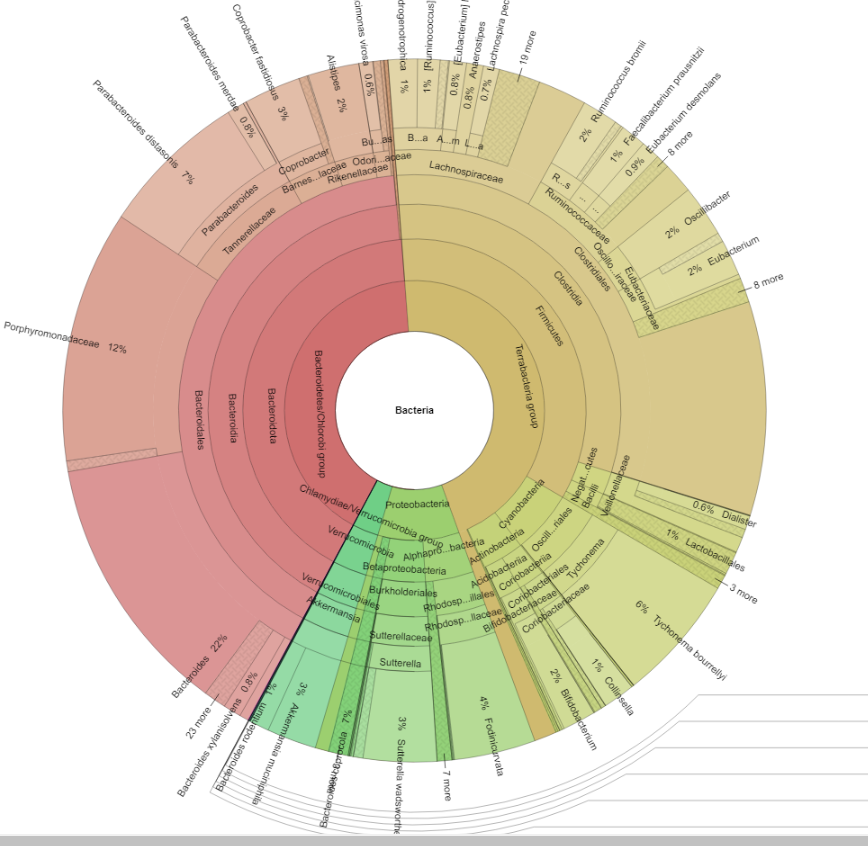
Where as a supported lab shows more details.
Recent Comments