For over a year I have made donated data available at: http://lassesen.com/ubiome/ . I would hope that anyone using open source software project to also be open data.
This post deals with exporting the taxon, continuous and category data to a csv file format suitable for importing to R or Python for data exploration. The program code is simple (with all of the work done in the shared library).
DataInterfaces.ConnectionString = "Server=LAPTOP-BQ764BQT;Database=MicrobiomeV2;Trusted_Connection=True; Connection Timeout=1000";
File.WriteAllLines("DataScience_Taxon.csv", DataInterfaces.GetFlatTaxonomy("species").ToCsvString());
File.WriteAllLines("DataScience_Continuous.csv", DataInterfaces.GetFlatContinuous( ).ToCsvString());
File.WriteAllLines("DataScience_Category.csv", DataInterfaces.GetFlatCategory().ToCsvString());
File.WriteAllLines("DataScience_LabReport.csv", DataInterfaces.GetLabReport().ToCsvString());
The result files examples are shown below:

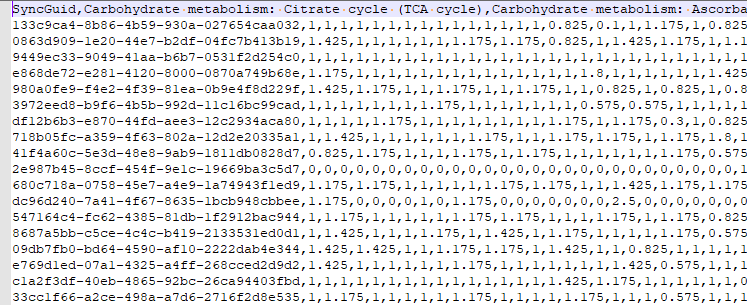
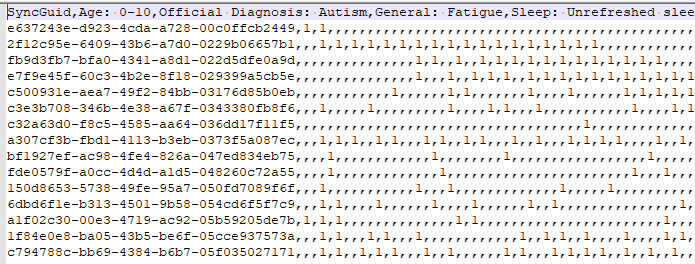
Source at:
Library update is shown below https://github.com/Lassesen/Microbiome2/tree/master/DataScienceExport
Recent Comments