We want to view relationships between microbiome and various medical conditions. There is also a need to know the impact of various probiotics on various conditions.
The International Classification of Disease (ICD) is the equivalent as taxon numbers for bacteria and is a well defined comprehensive list of medical conditions. We may need to add alternative names for some of the conditions to match common usage.
If something is not there (it should be), we can always add our own codes and diagnosis. For example there is no “SIBO” or ” Small intestine bacterial overgrowth” item, scanning on like we see that the code of A04.9 is often used. We would insert that into the ICDAltName table (likely two rows, one for each variation).
First we need to download some files as shown below.
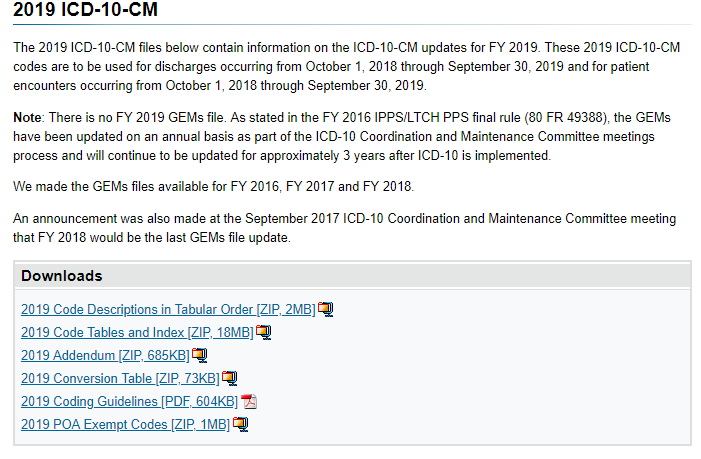
The file that we wish to use is shown below.
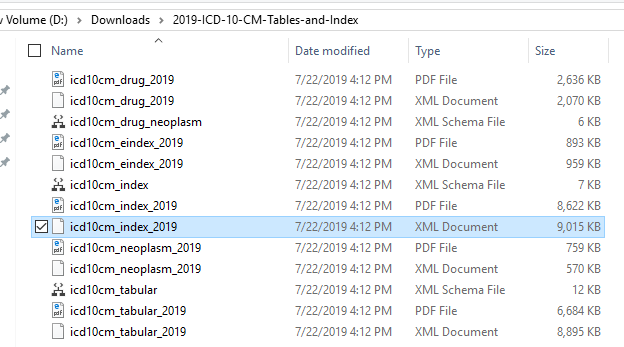
This file is the parameter to pass to the C# console application that will upload and populate the table. Most of the magic happens in SQL.
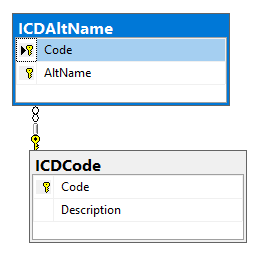
We end up with tables as shown below. In the next posts we look at connecting conditions to probiotics to studies, and conditions to microbiomes to studies.
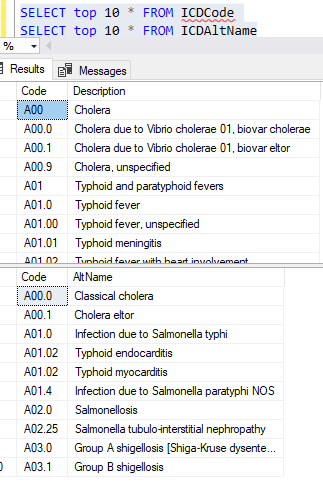
Example of the data after uploading is shown below. There are around 44 thousand IDC codes – fortunately, they are in a hierarchy.
The source code is at: https://github.com/Lassesen/Microbiome2/tree/master/UploadICD
You just need to give the location of the two files downloaded above
uploadICD.exe D:\Downloads\2019-ICD-10-CM-Tables-and-Index\icd10cm_index_2019.xml D:\Downloads\2019-ICD-10-CM-Tables-and-Index\icd10cm_tabular_2019.xml
Homework Item
We have some 44 thousand items which are in a hierarchy if you use the code. There are many ways to handle that on a web site — what approach will you do?
1 thought on “Importing medical conditions ( ICD tables)”
Comments are closed.